| Issue |
BIO Web Conf.
Volume 12, 2019
41st World Congress of Vine and Wine
|
|
|---|---|---|
| Article Number | 01019 | |
| Number of page(s) | 5 | |
| Section | Viticulture | |
| DOI | https://doi.org/10.1051/bioconf/20191201019 | |
| Published online | 19 February 2019 | |
A plant regeneration platform to apply new breeding techniques for improving disease resistance in grapevine rootstocks and cultivars
1 Department of Agricultural, Food and Environmental Sciences, Università Politecnica delle Marche, Ancona, Italy
2 Vitroplant International, Cesena, Italy
3 Vivaio Enotria, Treviso, Italy
Worldwide grapevine cultivation is based on the use of elite cultivars, in many cases strictly linked to local important wine brands. Most of Vitis viniferacultivars have high susceptibility to fungal and viral diseases therefore, new breeding techniques (e.g. Cisgenesis, RNAi and gene editing) offer the possibility to introduce new clones of the main cultivars with increased diseases resistance, in order to reduce environmental impact and improve quality in the intensive wine grape industry. This study is finalized to develop efficient in vitro regeneration and transformation protocols to extend the application of these technologies in wine grape cultivars and rootstocks. With this aim, in vitro regeneration protocols based on the production of meristematic bulks (Mezzetti et al., 2002) were optimized for different grapevine cultivars (Glera, Vermentino, Sangiovese, Thompson Seedless) and rootstocks (1103 Paulsen, and 110 Richter). The meristematic bulks were then used as explants for Agrobacteriummediated genetic transformation protocols, by comparing the use of NPTII and e-GFP as marker genes. Results confirmed the efficiency of meristematic bulks as the regenerating tissue to produce new modified plants in almost all the above genotypes. The highest regeneration efficiency in some genotypes allowed the selection of stable modified lines/calli with only the use of e-GFP marker gene. This protocol can be applied in the use of MYB marker gene for the production of cisgenic lines. Genotypes having the highest regeneration and transformation efficiency were also used for transformation experiments using a hairpin gene construct designed to silence the RNA-dependent RNA polymerase (RpRd) of the GFLV and GLRaV3, which would induce multiple virus resistances, and the Dicer-like protein 1 (Bc-DCL1) and Bc-DCL2 to control B. cinerea infection.
© The Authors, published by EDP Sciences, 2019
 This is an Open Access article distributed under the terms of the Creative Commons Attribution License 4.0 (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License 4.0 (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
1. Introduction
Grapes (Vitis spp.) are among the world's most important fruit crops for their diverse uses. Grapes are consumed fresh commonly referred to as table grapes, dried (sultanas, raisins) or as a high-value drink (juice, wine). However, grapevine cultivation is penalized by pathological problems with significant impact on production, quality and related costs. Traditional breeding techniques are limited in that there is reduced genetic resources and an increase in variability that is unacceptable for the preservation of traditional grapevine clones. Therefore, there is need to improve the quality and resistance to diseases through genetic improvements.
The application of New Breeding Techniques (NBTs), such as cisgenesis, RNAi and gene editing, can be a useful alternative to classical breeding methods to introduce or modify important agronomical traits in grapevine clones [1, 2]. The sequencing of V.vinifera genome has allowed the identification of gene sequences useful for controlling several important agronomical characteristics in grapevine, which has been mostly validated by Agrobacterium-mediated transformation [3–5]. The main bottleneck in the application of biotechnological tools for grapevine genetic improvement is the development of efficient and reproducible in vitro regeneration and transformation protocols, which often need to be optimized for the individual genotypes being studied [5]. In vitro grapevine regeneration has been achieved principally by somatic embryogenesis using different tissues as starting explants, such as anthers, leaves and ovaries [6–8]. In vitro grapevine shoots regeneration was also obtained via organogenesis, which represents a good alternative regeneration method to somatic embryogenesis, avoiding some technical problems related to embryogenetic cultures induction and maintenance [9–12].
Another important key point that is part of genetic engineering protocols applied to plant species, is the availability of an effective selective regime after infection of explants with Agrobacterium, that should lead to a rapid regeneration, which should only be restricted to the putative transgenic cells. In this sense, the combined use of antibiotic-based selectable marker genes and reporter genes, such as green fluorescent protein(GFP), represents an efficient selective strategy, which aids in avoiding the regeneration of chimeric tissues and/or escapes [13, 14]. In grapevine, it is really important to have efficient regeneration and transformation protocols for the most important cultivars and rootstocks for the application of NBTs aimed at achieving some of the main objectives such as, the induction of biotic resistance.
Grapevine is affected by numerous pathogenic diseases including viral diseases, such as “fanleaf (GFLV)” and “leaf roll (GLRaV)” diseases, which are the most diffused in Europe. The application of rigorous certification criteria is the only strategy available to control the diffusion of viruses. However, this method is not efficient in ensuring total control of these diseases thus; application of NBTs such as RNA interference (RNAi) has emerged as alternative tool to induce resistance to virus in several plant species. RNAi is a complex of cell mechanisms that evolved naturally in eukaryotes and used to degrade specific nucleic acid sequences and/or to inhibit gene expression [2, 15]. It has been mainly associated with a natural defence strategy against viruses and transposable elements in plants [16, 17]. This mechanism relies on the presence of non-coding double-stranded RNAs (dsRNAs) molecules, which are processed by the plant-cell machinery and cause the degradation, in a sequence-specific manner, of all the complementary viral transcripts present in the plant cell [17]. RNAi silencing signal is mobile and can move from cell to cell or for longer distance through the plant vascular system [15, 17]. This is of great interest when applied to rootstocks as a source of RNAi controlling plant virus infection into non-genetically modified scion.
In grapevine few examples exist of virus resistance induction by genetic engineering; indeed, the validation of RNAi-based gene constructs has been mainly carried out on model plants like Nicotiana benthamiana [18–23]. Recently it was discovered that a cross-kingdom RNAi mechanism exists, which is used by some plant and animals pathogens to silence host immunity genes [24]. An example is represented by the plant fungal pathogen B.cinerea, whereby mechanism of infection involves the introduction of small RNAs (sRNAs) able to suppress the plant cell immunity defences [25, 26]. Thanks to this discovery, resistance to B.cinerea was obtained in Arabidopsis and Tomato by expressing sRNAs that target B. cinerea Dicer-like protein 1 (Bc-DCL1) and Bc-DCL2; furthermore, gray mold disease was inhibited also by applying Bc-DCL1- and Bc-DCL2-derived sRNAs or dsRNAs molecules on the surface of fruits (tomato, strawberry and grape), vegetables (lettuce and onion), and flowers (rose) [27].
In the present study the regeneration protocol, via organogenesis, based on the induction of meristematic bulks [12], was optimized for different grapevine cultivars (Glera, Vermentino, Sangiovese, Thompson Seedless) and rootstocks (1103 Paulsen, and 110 Richter). To investigate the transformation efficiency of the different genotypes, the meristematic bulks created for each clone were used at the beginning as explants for Agrobacterium-mediated genetic transformation with a gene construct that expresses the enhanced green fluorescent protein (e-GFP) and neomycin phosphotransferase (nptii) as marker genes. The genotypes having the highest regeneration and transformation efficiency were used also to compare two selection strategies for the identification of putatively transformed tissues/shoots: i) the combined use of both nptii and eGFPmarker genes, ii) the visual screening based on the eGFP gene expression alone. The same genotypes were also exploited for transformation trials using two different hairpin gene constructs, one designed to silence the RNA-dependent RNA polymerase (RpRd) of the GFLV and GLRaV3, and another one that silence the Dicer-like protein 1 (Bc-DCL1) and Bc-DCL2 to control B. cinerea infection.
2. Materials and methods
2.1. Plant material
In vitro proliferated shoots of V.vinifera cultivars Thompson seedless, Vermentino, Glera, Sangiovese and the rootstocks 110 Richter (V.berlandieri x V.rupestris) and 1103 Paulsen (V.berlandieri x V.rupestris) were provided by Vitroplant Italia, and used as starting explants to induce meristematic bulks (MBs). The protocol described by Mezzetti et al., 2002 [12] was optimized for the single genotypes; it induces the production of MBs through the addition of increasing concentration of 6-benzylaminopurine (BAP) in the culture medium. The MBs obtained were cut into slices and exploited as vegetative material for the transformation trials (Fig. 1).
 |
Figure 1. Organogenesis-based regeneration method applied to grapevine: a) Grapevine Meristematic bulk; b) Slices obtained from meristematic bulk prepared for Agrobacterium-mediated transformation. |
2.2. Plasmid vectors and gene constructs for the transformation trials
Three different plasmids and gene constructs were used to achieve the transformation trials: i) the pK7WG2 binary vector expressing both eGFP and nptii genes under the control of the 35S Cauliflower mosaic virus (CaMV) promoter; ii) a derivative of pBin19 binary vector expressing the hairpin gene construct hp-GFLV-GLRaV, as described by Girolomini et al., 2012 [28], and iii) the phellsgate 8.0 containing the hairpin cassette designed to silence the Bc-DCL1 and Bc-DCL2 sequences [27]. All the gene constructs were inserted in A.tumefaciens strain EHA105.
2.3. Grapevine genetic transformation trials
All grapevine genotypes mentioned above were initially transformed with the gene construct expressing both eGFP and nptii marker genes to identify the most efficient clones in terms of transformation efficiency. The transformation protocol used by Mezzetti et al., 2002 [12] was applied to all the grapevine genotypes in this study. After A.tumefaciens infection all the explants were placed on regeneration medium (MS salt and vitamins [29], 30 g L−1 sucrose, 7 g L−1 plant agar, 13.2 μM BAP, 0.01 μM 1-Naphthaleneacetic acid (NAA)) supplemented with 70 mg L−1 kanamycin, and sub-cultured on fresh media every three weeks. A second transformation trial was carried out using the same gene construct (i.e 35S-eGFP-NPTII) to transform MBs of the genotypes that had shown the best results in term of transformation efficiency from the first transformation. Finally the putatively transformed lines were selected by comparing the use of nptii and e-GFP as marker genes, placing half of the infected explants on media containing kanamycin and half without it. For all transformation trials, data on the number of transformation events ((number of calli expressing e-GFP/total explants) × 100) were acquired, by observing the infected explant under the stereomicroscope Leica MZ10F, to detect the e-GFP fluorescing cells. Genotypes having the highest regeneration and transformation efficiency were also used as starting explants for transformation trials using the hairpin gene constructs hp-GFLV-GLRaV and hp-BcDCL1/DCL2. Identification of the putative transgenic lines was obtained by placing the infected explants on regeneration medium supplemented with 70 mg L−1 kanamycin, and sub-cultured on fresh media every three weeks. The selected explants were isolated and moved to rooting medium (MS salt and vitamins [29], 30 g L−1 sucrose, 7 g L−1 plant agar, 5 μM Indole-3-butyric acid (IBA), kanamycin 70 mg L−1).
3. Results and discussion
3.1. Induction of MBs from different grapevine cultivars and rootstocks
The in vitro regeneration protocol based on the production of meristematic bulks [12] was efficiently optimized for different grapevine cultivars (Glera, Sangiovese, Thompson Seedless, Vermentino) and rootstocks (1103 Paulsen, 110 Richter) (Fig. 2). This in vitro regeneration method, via organogenesis, has been confirmed, also by other studies, to be really effective for the regeneration and genetic transformation, not only of grapevine, but also of other plant species, as Prunus spp. and blueberry [30–33].
 |
Figure 2. Meristematic bulks of grapevine cultivars and rootstocks: a) Thomspon seedless; b) Sangiovese; c) Glera; d) Vermentino; e) 110 Richter; f) 1103 Paulsen. |
3.2. Genetic transformation and selection of putative transformed lines using 35S-eGFP-NPTII gene construct
The transformation trials were performed on a total of 100 explants per genotype. Although, all grapevine cultivars and rootstock showed high in vitro regeneration response, transformation competence presented differences between genotypes (Table 1). Thompson seedless and 110 Richter showed the highest transformation competence (among the cultivar and rootstocks under study), calculated as percentage of calli expressing e-GFP on total explants. Whereas, Glera showed the lowest transformation efficiency, by applying the protocol developed by us.
To compare the possibility of selecting transformed lines using a reporter gene (eGFP marker gene) alone, another transformation trial was performed only on MBs from Thompson seedless and 110 Richter. Half of the plant material was selected by both eGFP visual screening and kanamycin selection; the rest of plant explants were placed on the same medium but without the of addition kanamycin, and putative transgenic lines were identified through the detection of eGFP expression only. Results obtained showed that both methods of selection led to the obtainment of transgenic calli and/or shoots. In particular, transgenic shoots of Thompson seedless were isolated by both selection regimes (Fig. 3a–d), while only fluorescent putative transgenic proliferating calli were isolated from MBs of 110 Richter (Fig. 3e–h).
The combined use of e-GFP and nptii marker genes has shown some advantages for instance, a more rapid identification of putative transgenic lines, the reduction of escapes or chimeras regeneration (Fig. 3c, d, g, h). However, the introduction of antibiotic resistance into the plant genome presents some limits from a biosafety point of view and for the public acceptance [34]. The obtainment of transgenic calli/shoots selected only by the use of a reporter gene, demonstrates that this protocol can be applied in the use of grapevine-derived reporter gene system alone, for the production of cisgenic lines [35]. Nevertheless, the use of non-destructive reporter genes, like eGFP, has some main drawbacks, as they don't give a selective advantage to the transformed cells over the non-transformed ones hence, extending the time necessary for the obtainment of transgenic calli and/or shoots.
Transformation efficiency of different grapevine cultivars and rootstocks after 9 weeks of selection on kanamycin 70 mg L−1.
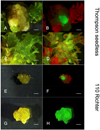 |
Figure 3. eGFP-fluorescing calli and/or shoots of Thompson seedless and 110 Richter after 9 weeks of culture on media without (a, b, e, f) or with (c, d, g, h) the addition of kanamycin in the medium. (bar = 2 mm). |
3.3. Introduction of hairpin gene constructs to induce Post-Transcriptional Gene Silencing (PTGS) in grapevine
A total of 200 MB slices of Thompson seedless and 110 Richter were used to perform transformation trials using a hairpin gene construct designed to silence both GFLV and GLRaV viruses [28]. Another set of transformation trials were carried out on a total of 200 MB slices of Thompson seedless to introduce the hairpin gene construct against the Bc-DCL1 and Bc-DCL2. The infected lines are currently under selection by kanamycin 70 mg L−1 and some putative transgenic lines have been isolated and transferred on rooting medium (Fig. 4) to be submitted to PCR and Southern blot analyses.
 |
Figure 4. Regeneration of putative transgenic lines of Thompson seedless infected with A.tumefaciens harbouring the hairpin construct Bc-DCL1/DCL2, after three months on selective culture medium a) and a putative transgenic line on elongation and rooting medium b). |
4. Conclusions
This study has confirmed the efficiency and adaptability of the in vitro regeneration protocol, developed by Mezzetti et al., 2002, for the obtainment of MBs of different cultivar and rootstock of Vitis, with a high shoot regeneration capability. The results obtained from the transformation trials, using the gene construct 35S-eGFP-NPTII, were useful as they helped to identify, among the genotype tested, Thompson seedless and 110 Richter as the best grapevine cultivar and rootstock, respectively, in terms of Agrobacterium-mediated transformation competence. These preliminary results show the possibility of using Thompson seedless as model genotype for future functional genomic studies, and for the validation of gene constructs, like RNAi-based ones. While, further studies need to be performed on the rootstock to improve its regeneration efficiency from putatively transformed cells; indeed only proliferating eGFP-fluorescent calli of 110 Richter have been obtained from these transformation trials. The use of eGFP gene as selectable marker alone led to the obtainment of transgenic events. The same method can be used in future for the selection of genetically modified plants, using grapevine-derived reporter genes, such as those belonging to the MYB gene family, so as to avoid the introduction of antibiotic-based selectable marker genes.
Acknowledgments
This work was supported by Vitroplant Italia, Enotria S.S. and Ampelos. We thank Prof. Tiziana Pandolfini and Dr. Barbara Molesini for providing the gene construct 35S-eGFP-NPTII and hairpin gene constructs against GFLV-GLRaV viruses. We thank Prof. Hailing Jin for providing the hairpin gene construct against Bc-DCL1/DCL2 sequences.
References
- D.J. Gray, Z.T. Li, S.A. Dhekney, Plant Sci. 228, 3 (2014) [CrossRef] [PubMed] [Google Scholar]
- C. Limera, S. Sabbadini, J.B. Sweet, B. Mezzetti, Frontiers Plant Sci. 8, 1418 (2017) [CrossRef] [Google Scholar]
- O. Jaillon, J.M. Aury, B. Noel, A. Policriti, C. Clepet, A. Casagrande, A. Vezzi, Nature 449, 463 (2007) [CrossRef] [PubMed] [Google Scholar]
- R. Velasco, A. Zharkikh, M. Troggio, D.A. Cartwright, A. Cestaro, D. Pruss, G. Malacarne, PloS one 2, e1326 (2007) [CrossRef] [PubMed] [Google Scholar]
- R. Saporta, S. Pedro-Galan, G. Domenech, M. Carmen, Vitis 55, 173 (2016) [Google Scholar]
- C.L. Rosenfield, S. Samuelian, J.R. Vidal, B.I. Reisch, Am. J. Enol. Vitic. 61, 348 (2010) [Google Scholar]
- Z.T Li, D.L. Hopkins, D.J. Gray, Transgenic Res. 24, 821 (2015) [CrossRef] [PubMed] [Google Scholar]
- L. Dai, Q. Zhou, R. Li, Y. Du, J. He, D. Wang, S. Cheng, J. Zhang, Y. Wang, Plant Cell Tiss. Org. Cult. 121, 397 (2015) [CrossRef] [Google Scholar]
- P. Iocco, T. Franks, M.R. Thomas, Transgenic Res. 10, 105 (2001) [CrossRef] [PubMed] [Google Scholar]
- K.L. Nicholson, N. Tarlyn, T. Armour, M.E. Swanson, A. Dhingra, PCTOC 111, 123 (2012) [CrossRef] [Google Scholar]
- M. Dutt, Z.T. Li, S.A. Dhekney, D.J. Gray, Plant Cell Reports 26, 2101 (2007) [CrossRef] [PubMed] [Google Scholar]
- B. Mezzetti, T. Pandolfini, O. Navacchi, L. Landi, BMC Biotechnol. 2, 18 (2002) [CrossRef] [PubMed] [Google Scholar]
- S.N. Maximova, A.M Dandekar, M.J. Guiltinan, Plant Mol. Biol. 37, 549 (1998) [CrossRef] [PubMed] [Google Scholar]
- A. Ballester, M. Cervera, L. Pena, Plant Cell Rep. 27, 1005 (2008) [Google Scholar]
- A.E.M. De Alba, E. Elvira-Matelot, H. Vaucheret, Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms 1829, 1300 (2013) [CrossRef] [Google Scholar]
- S.W. Ding, O. Voinnet, Cell 130, 413 (2007) [CrossRef] [PubMed] [Google Scholar]
- G.N.Y. Lemgo, S. Sabbadini, T. Pandolfini, B. Mezzetti, Transgenic Res. 22, 1073 (2013) [CrossRef] [PubMed] [Google Scholar]
- F. Maghuly, S. Leopold, A. da Câmara Machado, E.B. Fernandez, M.A. Khan, G. Gambino, M. Laimer, Plant Cell Rep. 25, 546 (2006) [Google Scholar]
- K.S. Ling, H.Y. Zhu, D. Gonsalves, Transgenic Res. 17, 733 (2008) [CrossRef] [PubMed] [Google Scholar]
- C. Monier, P. Barbier, B. Walter, In VII International Symposium on Grapevine Genetics and Breeding 528, 379 (1998) [Google Scholar]
- G. Gambino, I. Perrone, A. Carra, W. Chitarra, P. Boccacci, D. Torello Marinoni, M. Barberis, F. Maghuly, M. Laimer, I. Gribaudo, Transgenic Res. 19, 17 (2010) [CrossRef] [PubMed] [Google Scholar]
- F. Maghuly, S. Leopold, A.D.C. Machado, E.B. Fernandez, M.A. Khan, G. Gambino, I. Gribaudo, A. Schartl, M. Laimer, Plant Cell Reports 25, 546 (2006) [CrossRef] [PubMed] [Google Scholar]
- R. Jardak-Jamoussi, P. Winterhagen, B. Bouamama, C. Dubois, A. Mliki, T. Wetzel, G.M. Reustle, PCTOC 97, 187 (2009) [CrossRef] [Google Scholar]
- A. Weiberg, M. Wang, M. Bellinger, H. Jin, Annu. Rev. Phytopathol. 52, 495 (2014) [CrossRef] [PubMed] [Google Scholar]
- A. Weiberg, M. Wang, F.M. Lin, H. Zhao, Z. Zhang, I. Kaloshian, H.D. Huang, H. Jin, Science 342, 118 (2013) [Google Scholar]
- M. Wang, A. Weiberg, E. Jr. Dellota, D. Yamane, H. Jin, RNA Biol. 1, 8 (2017) [Google Scholar]
- M. Wang, A. Weiberg, F.M. Lin, B.P. Thomma, H. Huang, H. Jin, Nature Plants 2, 16151 (2016) [CrossRef] [PubMed] [Google Scholar]
- L. Girolomini, S. Sabbadini, B. Mezzetti, D. Palma, T. Pandolfini, A. Polverari, Acta Horticulturae 929, 393 (2012) [Google Scholar]
- T. Murashige, F. Skoog 15, 473 (1962) [Google Scholar]
- M. Pérez-Jiménez, A. Carrillo-Navarro, J. Cos-Terrer, Plant Cell Tissue Organ Cult. 108, 55 (2012) [Google Scholar]
- S. Sabbadini, T. Pandolfini, L. Girolomini, B. Molesini, O. Navacchi, Agrobacterium Protocols edited by K. Wang (Springer, New York, 2015), p. 205 [CrossRef] [Google Scholar]
- R. Cappelletti, S. Sabbadini, B. Mezzetti, Scientia Horticulturae 207, 117 (2016) [Google Scholar]
- X. Xie, C.B. Aguero, Y. Wang, M.A. Walker, Plant Cell, Tissue Organ Cult. 126, 541 (2016) [CrossRef] [Google Scholar]
- D. Breyer, L. Kopertekh, D. Reheul, Critical Rev. Plant Sci. 33, 286 (2014) [CrossRef] [Google Scholar]
- R. Kandel, D.R. Bergey, M. Dutt, V. Sitther, Z.T. Li, D.J. Gray, S.A. Dhekney, PCTOC 124, 599 (2016) [CrossRef] [Google Scholar]
All Tables
Transformation efficiency of different grapevine cultivars and rootstocks after 9 weeks of selection on kanamycin 70 mg L−1.
All Figures
 |
Figure 1. Organogenesis-based regeneration method applied to grapevine: a) Grapevine Meristematic bulk; b) Slices obtained from meristematic bulk prepared for Agrobacterium-mediated transformation. |
| In the text | |
 |
Figure 2. Meristematic bulks of grapevine cultivars and rootstocks: a) Thomspon seedless; b) Sangiovese; c) Glera; d) Vermentino; e) 110 Richter; f) 1103 Paulsen. |
| In the text | |
 |
Figure 3. eGFP-fluorescing calli and/or shoots of Thompson seedless and 110 Richter after 9 weeks of culture on media without (a, b, e, f) or with (c, d, g, h) the addition of kanamycin in the medium. (bar = 2 mm). |
| In the text | |
 |
Figure 4. Regeneration of putative transgenic lines of Thompson seedless infected with A.tumefaciens harbouring the hairpin construct Bc-DCL1/DCL2, after three months on selective culture medium a) and a putative transgenic line on elongation and rooting medium b). |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.



