| Issue |
BIO Web Conf.
Volume 25, 2020
International Scientific Online-Conference “Bioengineering in the Organization of Processes Concerning Breeding and Reproduction of Perennial Crops” 2020
|
|
|---|---|---|
| Article Number | 03001 | |
| Number of page(s) | 7 | |
| Section | DNA Technology in Producing New Varieties of Permanent Crops with Specified Features and Simplified Identification of Genotypes | |
| DOI | https://doi.org/10.1051/bioconf/20202503001 | |
| Published online | 01 October 2020 | |
Microsatellite DNA-markers in the study of the gene pool of fruit crops
Federal State Budget Scientific Institution «North Caucasian Federal Scientific Center of Horticulture, Viticulture, Wine-making», 39 str. 40 Let Pobedy, Krasnodar, 350901, Russia
* Corresponding author: supruni@mail.ru
This article describes development of multilocus SSR-markers sets for genotyping Pyrus, Prunus, and Malus from various genetic collections of the South of Russia. Generated multiplex sets of SSR-markers were used in the certification of cultivated varieties and in the analysis of the genetic structure of Pyrus, Prunus and Malus species collections. The results of SSR genotyping of pear, apple, plum and sweet cherry made it possible to establish genetic relationships between varieties, including groups of modern varieties of Russian and foreign breeding and, in turn, local autochthonous varieties. In general, the use of these multiplexes has confirmed their effectiveness in solving the assigned tasks.
© The Authors, published by EDP Sciences, 2020
 This is an Open Access article distributed under the terms of the Creative Commons Attribution License 4.0, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License 4.0, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
1 Introduction
At the present stage of development of selection and genetic research of the gene pool of cultivated plants, including fruit crops, special attention is paid to the use of methods that allows in a short time and with a high level of accuracy to characterize the studied gene pool. This concerns both methods for assessing a complex of phenotypic traits and methods that allows to assess genotypic variability. DNA marker analysis, based on the use of molecular genetic methods, makes it possible to effectively characterize the genotypes of the studied plant objects both breeding forms and samples from collections of genetic resources. The main areas of application of molecular genetic methods when working with genetic resources can be defined as the following: mapping and identification of genes that determine economically valuable traits, DNA-fingerprinting and certification of genotypes (varieties, hybrids), study of genetic relationships between samples/groups of samples, and also the study of the genetic structure of the gene pool of the species, the formation of so-called core collections, including the most genetically heterogeneous samples from main collection.
Among the methods based on the use of DNA marker technologies, one of the most informative is the method based on the analysis of polymorphism of microsatellite genome sequences. Their advantages include codominance, multi-allele, locus-specificity, distribution throughout the genome, and a high level of reproducibility of results. This determines the relevance and high prospects of their use in the DNA certification of the gene pool and the study of genetic diversity. Over the past ten years, a significant amount of research has been carried out on the gene pools of the genera Pyrus, Malus, and Prunus using SSR-markers.
Thus, the genetic collection of Italian apple varieties of 418 apple genotypes, including 383 samples of Italian selection and 35 world varieties, was analyzed using 15 SSRs [1]. Also, 16 SSR-markers were used to assess the genetic diversity of 87 apple samples from three Portuguese genetic collections [2]. Along with the data on the genetic structure of the collections, SSR-fingerprints of the studied cultivars were obtained. In a larger study, 16 SSR-markers were used to study 391 samples from the Chinese collection of the Academy of Agricultural Sciences, including samples of the species Malus x domestica Borkh., as well as its wild relatives Malus baccata, Malus prunifolia, Malus robusta, and Malus sieversii. The study made it possible to suggest that the M. domestica varieties from the former Soviet republics are most closely related to M. sieversii and may represent an independent line of domesticated apples, different from the varieties of Western Europe and North America [3]. Research using SSR markers within the Malus genus has not been limited to cultural forms. In particular, a set of SSR markers was used to study the genetic diversity and population structure of wild representatives of the genus Malus, including the Himalayan apple trees [M. baccata (L.) Borkh. and M. sikkimensis (Wenzig) Koehne ex C. Schneider] [4]. Another study used SSRs to assess the genetic relationships between 15 biotypes of Malus prunifolia. Data clustering showed that some of the grouped biotypes largely coincided in geographic distribution [5].
SSR-markers have also been widely used in genetic studies of members of the Pyrus genus. A team of researchers analyzed the genotypes of Hungarian pear varieties and developed a protocol for molecular identification and interpretation of data on varieties [6]. The national Portuguese genetic collections have a wide variety of traditional pear varieties. Eleven markers were used to assess the genetic diversity of 130 local varieties [7]. The aim of the research team was to determine the taxonomic relationship between pear varieties using the 7 SSRs. The study was carried out on the genotypes of twenty-six pears from Europe (Pyrus spinosa Forssk., Pyrus communis L., Pyrus elaeagrifolia Pall., Pyrus nivalis Jacq.), 18 Asian Pyrus pyrifolia (Burm.f.) Nakai pears and 4 of their hybrids (P communis × P. pyrifolia) [8]. The genetic diversity of 478 Pyrus accessions, including ancient Chinese cultivars, breeding cultivars and wild accessions, as well as introduced pear cultivars from Japan and Korea, were studied using a set of 17 SSRs markers distributed across all 17 linkage groups of the pear genome [9].
Among the recent works on genotyping Prunus collections based on microsatellite polymorphism, a number of studies can be distinguished. In 2020, data were published on the successful testing of EST-SSR on the peach gene pool [10], and in the current year, the results of evaluating 104 samples of domestic plum from various European collections [11] were published. In 2019, a group of researchers reported on the results of genotyping of local plum species in Tunisia, genetic diversity was assessed by the polymorphism of microsatellite loci and the S-gene [12], another research team in a 2019 article outlined the results of a genetic study of almonds from various ecological-geographical regions [13]. It is worth noting a number of studies using SSR markers carried out on sweet cherry genotypes and cherry species, including the work of Russian scientists [14, 15, 16].
It is obvious that microsatellite markers represent an important genetic tool in the study of the genetic diversity of fruit crops. In this regard, given the high relevance of research in this area in the world, as evidenced by a significant number of scientific publications in international rating journals, we set the task of studying the genetic diversity and DNA-certification of the gene pool of fruit crops in southern Russia using microsatellite DNA-markers.
In accordance with the goal, the tasks of our research include:
- improving the methodology of DNA-marker certification of fruit crops through the use of multiplex SSR-analysis;
- performing DNA-certification based on the analysis of microsatellite loci and studying the genetic structure of the gene pool of fruit crops.
2 Materials and methods
The object of the study is autochthonous and modern varieties of apple, pear, sweet cherry, peach, apricot, cherry plum, domestic plum from the collections of genetic resources of the South of Russia, as well as species samples and interspecific hybrids of the genera Malus, Prunus, Pyrus. DNA extraction is carried out using the CTAB method with minor modifications [1]. PCR according to the following program: denaturation for 3 minutes at 94 °C; then 35 cycles: 10 seconds at 94 °С denaturation, 45 seconds at Ta primer annealing, 45 seconds at 72 °С elongation; the final elongation cycle is 5 minutes at 72 °C. The primer annealing temperature (Ta) varied depending on the primer. The following standardized concentrations of the components of the reaction mixture were used: 0.05 mM deoxynucleotide triphosphates, 0.3 μM each primer, 2.5 μl PCR buffer, and 1 unit. Taq DNA polymerase (SibEnzyme LLC), 50 ng DNA. PCR was performed in a total volume of 25 μl. A preliminary analysis of the amplification quality was carried out using electrophoresis in 2% agarose or 8% polyacrylamide gel. The analysis of the size of the amplified fragments was carried out using an automatic genetic analyzer ABIprism 3130.
3 Results and discussion
At the stage of selection of DNA markers for research, the following parameters were taken into account:
- the level of polymorphism in the study of foreign gene plasma of target species, in accordance with data from the world scientific literature;
- the level of reproducibility (the possibility of obtaining reliably reproducible analysis results) in different species within the genus (for SSR markers of the genus Prunus) and between genera (for the genera Malus and Pyrus);
- the degree of use of DNA markers in the study of the world gene pool of target species (the priority in the selection was the DNA markers with the highest degree of use in the research of the world gene pool of fruit plants);
- primer annealing temperature and size of synthesized PCR fragments for more efficient formation of multiplex sets;
Figure 1 shows the results of testing the microsatellite marker CH03d07 (the results of electrophoresis of amplification products in 8% polyacrylamide gel are shown).
The main task in optimizing PCR parameters was to obtain maximum synthesis of target fragments with minimum synthesis of nonspecific amplificates. In addition, in order to reduce the time spent in performing genotyping of a larger number of samples and to simplify the algorithm of work, one or two unified PCR protocols were developed for all SSR markers used in the work and, accordingly, for all multiplex sets. As a result, for most of the SSR markers used in the work, the most optimal PCR program was adopted, which is given in the section “Material and research methods”.
The used DNA markers were grouped into multiplex sets according to the size of the amplified fragments in such a way that SSR markers with non-overlapping ranges of amplified sequence sizes were included in each set. This is necessary to prevent overlapping of the fluorescence spectra of amplification fragments when analyzing their sizes using capillary electrophoresis on the ABIprism3130 genetic analyzer.
M – DNA-ladder (molecular weight marker); apple varieties: 1-Sun, 2-Kurnakovskoe, 3-Secret, 4Zephyr, 5-Talida, 6-Lyubava, 7-Feya, 8-Ekaterinodarskoe, 9-Nocturne, 10-Talisman, 11-Orlovskoe woodlands, 12-Soyuz, 13-Start, 14-Aphrodite, 15-Imrus, 16-Prima
For each marker in the multiplex set, a specific fluorescent dye was used: carboxyfluorescein (FAM), 6-carboxyrodamine (R6G), carboxy-X-rhodamine (ROX), tetramethylcarboxyrodamine (TAMRA). Each of these fluorophores has different optical spectra, which makes it possible to simultaneously identify several fragments synthesized using primers labeled with different dyes in one reaction. Figures 2 and 3 show the results of a fragmentary analysis of the studied samples of fruit crops using some multiplex sets of SSR markers formed during the research. The results are presented in the working window of the GeneMapper 4.1 program.
Two peaks / fragments for markers CPSCT004 in the peach cultivar Ranniy Kubani and BPPCT028 in both cultivars indicate the heterozygosity of microsatellite loci, the presence of only one fragment (CPSCT004 in cultivar Pamyat Simirenko, BPPCT007 in both cultivars) indicates homozygosity. Locus BPPCT037 is heterozygous in the presented varieties, loci UDP98-412 and BPPCT023 are homozygous.
The presence of two peaks/two fragments of the same color on the electrophoregram (UDP98-022, EMPaS06) indicates heterozygosity of microsatellite loci, the presence of only one fragment (UDP96-018) indicates homozygosity.
In the course of research carried out from 2011-2012, a total of about 40 multiplex sets were formed. The number of SSR-markers included in these sets is as follows: for the genus Malus - 26 markers / 8 multiplexes; for the genus Pyrus - 23 markers / 7 multiplexes; for the genus Prunus - 93 markers / 25 multiplexes. At the moment, microsatellite genotyping of significant samples of samples has been carried out: apple tree - about 530 samples; pear - about 300 samples; stone fruit crops (including peach, apricot, sweet cherry, house plum, cherry plum) about 400 samples.
The genotyping results are accumulated in the working database for the accumulation of SSR genotyping results. A print screen of a part of the database is shown in Figure 4. The database contains the size of the amplified fragments identified for each test sample for a specific marker. For each SSR-marker, the color used is the fluorochrome used.
At the moment, according to the results of SSR genotyping for two fruit crops (apple and sweet cherry), the state registration of databases of DNA fingerprints of varieties has been completed: №2015620372 “Databank of DNA fingerprints of apple varieties based on the results of analysis of polymorphism of microsatellite genome loci”; # 2016620116 “Database of SSR-fingerprints of sweet cherry varieties”. Similar databases for pear and peach are in the process of registration.
Based on SSR genotyping for crops - objects of research, the study of genetic relationships, including between groups of modern varieties of both Russian and foreign breeding and local autochthonous varieties, was carried out. Data on close genetic relationships of autochthonous pear varieties of the North Caucasus and samples of Caucasian pears, selected in natural populations, were obtained. This fact testifies in favor of the fact that the selection of autochthonous pear varieties on the territory of the North Caucasus was carried out, first of all, through selection from local populations. This fact indicates the existence of an independent center for pear domestication in the North Caucasus [17]. When studying the genetic relationships of modern apple varieties and autochthonous varieties of southern Russia, complex genetic relationships were revealed between the autochthonous varieties of the Crimea and the Caucasus. Analysis of the structure of the collection of the sweet cherry gene pool showed a high level of genetic relationship between some varieties of Ukrainian selection and varieties of the selection of the FSBSI NCFSCHVW. This is consistent with the fact that Ukrainian varieties were widely involved in the breeding process at the research center. The study carried out on the domestic plum made it possible to establish the intervarietal relationship of the studied samples, as well as to identify two main groups from which the gene pool of the species was formed: thorns and cultivars [18].
 |
Fig. 1. Testing of SSR DNA marker CH03d07 (visualization in 8% polyacrylamide gel). |
 |
Fig. 2. Results of fragment analysis of peach cultivars Ranniy Kubani (A) and Pamyat Simirenko (B) using a multiplex set including SSR-markers CPSCT004, BPPCT007, BPPCT028. |
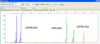 |
Fig. 3. The results of the fragment analysis of the Krasnodarskaya ranniaya cherry cultivar based on the multiplex set, including SSR-markers UDP98-022, EMPaS06, UDP96-018. |
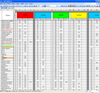 |
Fig. 4. Printscreen of SSR-fingerprint data accumulation database. |
4 Conclusion
Thus, based on the above, we have adapted the SSR-marking methods for genetic analysis of the gene pools of fruit crops in the South of Russia. From the SSR-markers selected in the literature, multiplex sets were formed to work within individual genera Pyrus, Malus, and Prunus. In total, we selected approximately 40 multiplex kits for the genera Malus, Pyrus and Prunus, a total of 142 markers were used to create multiplexes. The markers formed in multiplex sets made it possible to carry out mass genotyping of the collections of the fruit crops most demanded in the South of Russia. The carried out DNA certification made it possible to form databases of DNA fingerprints. In the future, these databases will be involved in work on varietal identification. On the other hand, the developed multiplex sets were used to assess the genetic structure of collections of fruit crops and related wild species in the South of Russia. The analysis of fruit gene pools carried out by the SSR made it possible to establish related ties between varieties in various crops of cherry, apple, plum and pear. Thus, the data obtained on the basis of the use of SSR-markers made it possible to establish family ties between the gene pools of the Ukrainian and Kuban cherries, reveal the isolated position of the autochthonous pear varieties of the North Caucasus relative to the world gene plasma, determine the genetic structure of the domestic plum collection and clarify the complex genetic relationships between the autochthonous apple varieties of the Crimea and the Caucasus. The obtained information will allow in the future to more effectively use the genetic potential of fruit collections for breeding purposes.
References
- W. Liang, L. Dondini, P. De Franceschi, R. Paris, S. Sansavini, S. Tartarini, Plant Mol. Biol. Report., 33(3), 458–473 (2015). https://doi.org/10.1007/s11105-014-0754-9 [Google Scholar]
- V. Ferreira, A. M. Ramos-Cabrer, V. Carnide, O. Pinto-Carnide, A. Assunção, A. Marreiros, R. Rodrigues, S. Pereira-Lorenzo, I. Castro, Tree Genet. Genomes, 12(3), 36 (2016). https://doi.org/10.1007/s11295-016-0997-8 [Google Scholar]
- Y. Gao, F. Liu, K. Wang, D. Wang, X. Gong, L. Liu, C. M. Richards, A. D. Henk, G.M. Volk, Tree Genet. Genomes, 11(5), 106 (2015). https://doi.org/10.1007/s11295-015-0913-7 [Google Scholar]
- C. Kumar, S. K. Singh, R. Singh, K. K. Pramanick, M. K. Verma, M. Srivastav, G. Tiwari, D.R. Choudhury, Genet. Resour. Crop Evol., 66, 1311–1326 (2019). https://doi.org/10.1007/s10722-019-00780-y [Google Scholar]
- M. Fu, F. Ma, Sci. Hortic., 146, 169–174 (2012). https://doi.org/10.1016/j.scienta.2012.08.020 [CrossRef] [Google Scholar]
- G. M. Kocsisné, D. Bolla, U. C. M. Anhalt-Brüderl A. Forneck, J. Taller, L. Kocsis, Genet. Resour. Crop Evol. (2020. https://doi.org/10.1007/s10722-020-00937-0 [PubMed] [Google Scholar]
- A. Queiroz, J. B. Guimaraes, C. Sanchez, F. Simoes, R. M. de Sousa, W. Viegas, M.M. Veloso, Sustainability, 11(19), 5340 (2019). [Google Scholar]
- Y. K. Kim, K. H. Won, U. Y. Lee, S. H. Yim, I. S. Shin, S. S. Kang, J. D. Han, H. C. Lee, Acta Hortic., 1094, 67–73 (2015). https://doi.org/10.17660/ActaHortic.2015.1094.5 [Google Scholar]
- L. Xue, Q. Liu, H. Hu, Y. Song, J. Fan, B. Bai, M. Zhang, R. Wang, M. Qin, X. Li, J Wu, Tree Genet. Genomes, 14, 48 (2018). https://doi.org/10.1007/s11295-018-1255-z [Google Scholar]
- M. K. Dehkordi, S. Beigzadeh, K. Sorkheh, J. For. Res., 31, 1359-1370 (2020). https://doi.org/10.1007/s11676-019-00922-z [Google Scholar]
- F. Gaši, J. Sehic, J. Grahic, S. H. Hjeltnes, M. Ordidge, D. Benedikova, M. Blouin-Delmas, P. Drogoudi, D. Giovannini, M. Hofer, K. Kahu, S. Kovacs, G. L. cis, M. Lateur, T. B. Toldam-Andersen, V. Ognjanov, H. Nybom, Genet. Resour. Crop Evol., 67, 1137-1161 (2020). https://doi.org/10.1007/s10722-020-00901-y [Google Scholar]
- G. Baraket, D. Abdallah, S.B. Mustapha, H. B. Tamarzizt, A. Salhi-Hannachi, Biochem. Genet., 57, 673–694 (2019). https://doi.org/10.1007/s10528-019-09922-4 [CrossRef] [PubMed] [Google Scholar]
- J. Halász, O. Kodad, G. M. Galiba, I. Skola, S. Ercisli, G. A. Ledbetter, A. Hegedűs, Tree Genet. Genomes, 15, 12 (2019). https://doi.org/10.1007/s11295-019-1319-8 [Google Scholar]
- A. Farsad, M. Esna-Ashari, Cytol. Genet., 50, 8–19 (2016). https://doi.org/10.3103/S0095452716010059 [CrossRef] [PubMed] [Google Scholar]
- J. A. Campoy, E. Lerigoleur-Balsemin, H. Christmann, R. Beauvieux, N. Girollet, J. Quero-García, E. Dirlewanger, T. Barreneche, BMC Plant Biol., 16, 49 (2016). https://doi.org/10.1186/s12870-016-0712-9 [CrossRef] [PubMed] [Google Scholar]
- A. Khadivi-Khub, Z. Zamani, R. Fattahi, A. Wünsch, Trees, 28, 471–485 (2014). https://doi.org/10.1007/s00468-013-0964-z [CrossRef] [Google Scholar]
- I. I. Suprun, S. V. Tokmakov, I. A. Bandurko, E. T. Ilnitskayaa, Russ. J. Genet., 52(11), 1149–1156 (2016). https://doi.org/10.1134/S1022795416100112 [Google Scholar]
- I. I. Suprun, I. V. Stepanov, S. V. Tokmakov, G. V. Eremin, Russ. J. Genet., 55(2), 172–179 (2019). https://doi.org/10.1134/S1022795419010149 [Google Scholar]
All Figures
 |
Fig. 1. Testing of SSR DNA marker CH03d07 (visualization in 8% polyacrylamide gel). |
| In the text | |
 |
Fig. 2. Results of fragment analysis of peach cultivars Ranniy Kubani (A) and Pamyat Simirenko (B) using a multiplex set including SSR-markers CPSCT004, BPPCT007, BPPCT028. |
| In the text | |
 |
Fig. 3. The results of the fragment analysis of the Krasnodarskaya ranniaya cherry cultivar based on the multiplex set, including SSR-markers UDP98-022, EMPaS06, UDP96-018. |
| In the text | |
 |
Fig. 4. Printscreen of SSR-fingerprint data accumulation database. |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.



