| Issue |
BIO Web Conf.
Volume 23, 2020
II International Scientific Conference “Plants and Microbes: The Future of Biotechnology” (PLAMIC2020)
|
|
|---|---|---|
| Article Number | 03005 | |
| Number of page(s) | 7 | |
| Section | Plant-Microbe Symbiosis, Including Natural and Artificial Symbiotic Systems | |
| DOI | https://doi.org/10.1051/bioconf/20202303005 | |
| Published online | 14 August 2020 | |
Studying the effect of tissue-specific expression of the K1 gene encoding LysM-receptor-like kinase on the development of symbiosis in peas
All-Russia Research Institute for Agricultural Microbiology, Laboratory of signal regulation, 196608 Saint-Petersburg, Russia
* Corresponding author: dol2helen@yahoo.com
To study the role of pea LysM receptor-like kinase K1 in the coordination of the infection process, starting in epidermis and nodule organogenesis in the root cortex of plants, during the development of rhizobium-legume symbiosis, the genetic constructs in which K1 gene was cloned under the control of tissue-specific promoter pLeEXT1 of tomato Lycopersicon esculentum extensin gene and the constitutive promoter of cauliflower mosaic virus (CaMV35S, cauliflower mosaic virus 35S) were obtained. During the transformation of the Nod- mutant line, the k1-1, with two types of constructs, the restoration of nodule formation was observed, which indicated the possible participation of K1 in the control not only early, but also later stages of symbiosis development in pea.
© The Authors, published by EDP Sciences, 2020
 This is an Open Access article distributed under the terms of the Creative Commons Attribution License 4.0, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License 4.0, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
1 Introduction
The formation of the symbiosis between legumes and nodule bacteria leads to the development of nitrogen-fixing nodules on the roots of plants. Genetic analysis of legume mutants impaired at subsequent stages of symbiosis development has shown that the process of forming functional nodules can be associated with the implementation of two main programs – infection process starting in the epidermis cells and nodule organogenesis in the root cortex[1–4]. These programs are developed individually, but in a way that depends on each other [4-6]. Binding of rhizobial signal molecules Nod factors with LysM-containing receptor-like kinases (LysM-RLK), localized in the plasma membrane of epidermis cells (mainly root hairs), leads to activation of the program that provides the penetration of rhizobia into plant root cells. This is accompanied by curling of root hairs, the formation of a “pocket” in which the microcolony of rhizobia is developed. Changing the structure of the cell wall under the influence of lytic enzymes leads to invagination of the epidermal cell membrane and the gradual formation of an infection thread (IT), by means of which rhizobia move through the cells of the epidermis to the root cortex. Simultaneously with the growth of IT in the epidermis, dedifferentiation and activation of cell divisions of the root cortex, which give rise to nodule primordia [7–10]. The implementation of two programs is completed by the penetration of IT into the cells of the root cortex of plants, the release of bacteria from ITs, the formation of symbiosomes and the initiation of the nitrogen fixation process in nodules.
In recent years, receptors to Nod factors and the main regulators important for signal transduction in epidermis cells have been identified and characterized [11-12]. In addition, transcription factors and regulators of plant response to the action of hormones cytokinins and auxins, which are necessary for the nodule organogenesis program activation in legumes, were identified [13–15]. However, how the interaction between the two programs related to different tissues (the epidermis and the cortex) is carried out remains unclear.
A convenient approach for studying the function of various genes in controlling the symbiosis development can be the transformation of plants using tissue-specific promoters that provide the activation of genes only in certain tissues. To study the effect of tissue-specific expression, a promoter of the tomato Lycopersicon esculentum EXT1 gene encoding an extensin-like cell wall protein can be used [16]. Analysis using reporter constructs allowed us to show the specific localization of gene expression in root hair cells (trichoblasts) during the transformation of tomato, potato and tobacco. The pLeEXT1 promoter was also used in the transformation of legumes, in particular, Medicago truncatula, to study the localization of the DsRED-E5 reporter gene. Expression of the reporter gene was also associated with the root hairs of M. truncatula and occurred in response to inoculation by rhizobia under the influence of ethylene [17-18]. Thus, obtaining genetic constructs under the pLeEXT1 promoter can be a convenient tool for studying the influence of selective gene expression in the epidermis upon the development of symbiosis in legumes.
Previously, we described the role of LysM-RLK K1 in controlling the symbiosis development in pea P. sativum L. Analysis of mutants impaired in this gene, as well as a number of other evidences, suggested that K1 can control both early symbiotic reactions (deformations and curling of root hairs) and later ones associated with the development of IT in the root cortex of plants and the release of bacteria from IT [12, 19]. Studying the effect of expression of this gene in the epidermis under the pLeEXT1 promoter on events occurring in the root cortex may be a convenient tool for identifying the relationship between the developmental programs in different types of tissues in pea during symbiosis.
In this work, we performed experiments on the complementation of the pea mutant k1-1 using the pLeEXT1::K1 construct containing the K1 gene under the promoter of the tomato extensin gene. Composite plants were obtained by transformation by Agrobacterium rhizogenes. The k1-1 mutant line was chosen by us as a model for studying the features of the tissue-specific pLeEXT1 promoter in pea plants, since it almost completely lacks responses to inoculation by rhizobia. The comparative analysis was performed using the p35S::K1 construct, which was previously used to carry out complementation and restore the function of nodule formation in the mutant k1-1 pea line.
2 Materials and methods
2.1 Strains and plants
Escherichia coli TOP10 strain (Thermo Fisher Scientific, USA) was used for standard cloning procedures. Strains of A. rhizogenes Arqua1 were used for plant transformation. Pea plants were inoculated with Rhizobium leguminosarum biovar viciae strain CIAM 1026 (WDCM 966) labelled with GUS [12]. We used P. sativum L. mutant line k1-1 and its parental line Cameor [12].
2.2 Genetic constructs obtaining
The pDONR P4-P1 vector containing the pLeEXT1 promoter (1121 p. o.) of the extensin gene was kindly provided by Dr. Sandra Bensmihen (INRA, Toulouse, France). We also used the pENTRY P4-P1 vector (Ghent University, Belgium) containing the 35S promoter of CaMV. Constructs with promoters were used for multi-locus homologous recombination with the pENTRY/D-TOPO P1-P2R vector containing the encoding sequence of the K1 gene, as well as pENTRY P2-P3R containing the t35s terminator (Ghent University, Belgium). The constructs were obtained using LR clonase II (Thermo Fisher Scientific, USA). pKm43GW was used as the final vector for cloning based on the homologous recombination [20]. The constructs were introduced into the A. rhizogenes Arqua1 strain by electroporation.
2.3 Plant transformation and histochemical analysis
The transformation of plants was performed using the method described earlier [21]. Histochemical staining of roots was performed using the method described earlier [12].
3 Results and discussion
In pea plants, infection of epidermis tissues begins in about 24-48 hours after inoculation, and nodule organogenesis is activated later (the first cell divisions of the pericycle and inner cortex can be detected on the 5th day after inoculation) [22]. Due to the fact that Nod factors do not penetrate into the root tissues of the plant and remain associated with receptors on the surface of root hairs, it is of interest to find out how the IT development program in the root cortex and nodule organogenesis are remotely activated.
In order to study the effect of tissue-specific expression of the K1 gene encoding the receptor to Nod factors on the development of early symbiotic reactions in peas and the formation of nodules, two constructs were created in which the K1 gene was under the p35S or pLeEXT1 promoters. The resulting vectors were named pKm43GW p35S::K1::t35S and pKm43GW pLeEXT1::K1::t35S, respectively. The design schemes are shown in Figure 1 (Fig. 1). The obtained constructs were used for transformation of the k1-1 pea mutant line using the A. rhizogenes Arqua1 strain.
In the k1-1 mutant line, no responses to rhizobial inoculation were observed, since the function of K1 receptor to Nod factors was impaired [12]. After transformation of the k1-1 mutant line with two types of constructs - pKm43GW p35S::K1::t35S and pKm43GW pLeEXT1::K1::t35S – the effective nodule formation was restored on transgenic roots (Fig. 2). The analysis of the deformations and curling of root hairs, as well as nodule primordia and nodules themselves was performed on 14 days after inoculation with rhizobia. The number of nodules and primordia was comparable when using two types of structures (Fig. 3). Thus, the program for the formation of infected nodules in peas.
Previously, the use of the pLeEXT1 promoter in the model legume M. truncatula, which has a similar type of nodule formation with pea, showed the possibility of using this promoter to study the specific expression of genes encoding the receptors to Nod factors that control the symbiosis development, such as NFP [23]. The nfp mutant of M. truncatula, as well as the pea k1-1 mutant, is completely not susceptible to rhizobial infection (Nod- phenotype). The NFP receptor does not have an active kinase domain and, probably, in combination with another receptor-like kinase, activates signal transduction in response to Nod factor binding. In peas, the orthologous to NFP, the Sym10 gene, encodes the receptor forming pair with LysM-RLK K1 during the initiation of symbiosis [12, 24]. Experiments on localization of NFP gene expression have shown that it is activated both in the epidermis and in the root cortex of M. truncatula during symbiosis [23, 25]. When NFP was synthesized only in epidermal cells under the control of the pLeEXT1 promoter, deformations and curling of root hairs were observed in the nfp mutant, but the ITs failed to develop [23]. At the same time, in the transformed pLeEXT1:: NFP plants, the cortical cell divisions are activated and nodule-like structures are appeared on the roots, which remained uninfected. In contrast, in nfp mutant transformed with construct under the constitutive promoter p35S::NFP, the complete recovery of nodule formation was observed. The authors suggested that local synthesis of NFP under the control of the pLeEXT1 promoter in epidermis cells is not sufficient to generate a signal that is necessary to start the IT growth and its progression through the cells of the root cortex [23, 26]. However, activation of NFP in the epidermis is sufficient to transmit a signal into the root cortex and stimulate cell division, that leads to the appearance of nodule-like structures. A possible explanation for these results may be the presence of several complexes of receptors to Nod factors in the epidermis, one of which is necessary for the initiation of the earliest stages of symbiosis – deformations of root hairs and reactivation of cell divisions in the root cortex (complex I), while the other complex works at later stage and controls the distribution of IT through the root cortex (complex II). Probably, NFP is not included in this complex II in M. truncatula plants. In contrast, as we showed the LysM-RLK K1 can completely restored the nodule formation during synthesis under the epidermal promoter.
 |
Fig. 1 Scheme of pKm43GW p35S::K1::t35S (a) and pKm43GW pLeEXT1::K1::t35S (b) vectors for pea plant transformation. |
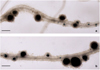 |
Fig. 2 Nodules on transgenic roots of k1-1 pea mutant line transformed with vectors a) p35S::K1::t35S b) pLeEXT1::K1::t35S. Plants were inoculated with GUS-labeled R. leguminosarum bv. viciae CIAM1026. Scale bar – 0, 3 mm. |
4 Conclusions
In our work, we investigated the possibility of the LysM-RLK K1 activation using the epidermal pLeEXT1 promoter in a k1-1 pea mutant, which has almost no reaction to rhizobial inoculation. Both deformations and curling of root hairs as well as the nodule formation infected with bacteria were observed on transgenic roots. Similarly, when the K1 gene was introduced under the p35S constitutive promoter, the k1-1 mutant restored its ability to form effective nodules, that corresponded to our previous results [12].
The results obtained in this work confirmed our earlier conclusions based on the phenotypic analysis of mutants impaired in the k1 gene, that LysM-RLK K1 can be involved not only in the initiation of symbiosis development, but also in the control of IT progress through the root cortex cells to the developing nodule primordia, as well as the release of bacteria into plant cells [12]. Further experiments will allow us to find out how K1 may regulate both of these processes – whether the receptor is re-localized in developing IT to control later stages of symbiosis development, or whether additional synthesis of the receptor is necessary in the root cortex tissues to regulate the infection.
The research was supported by a grant from the Russian Science Foundation (project no. 16-16-10043).
 |
Fig. 3 The number of nodules and primordia on the transgenic roots of the k1-1 line transformed with the p35S::K1::t35S and pLeEXT1::K1::t35S constructs during inoculation with GUS-labelled R. leguminosarum bv. viciae strain. The data of 4 independent biological repeats were analyzed. The Mann-Whitney U-test was used to estimate the differences between two variants. The differences between variants can be considered as insignificant (using significance level p = 0.05). |
References
- G.E.D. Oldroyd, J.A. Downie, Annu. Rev. Plant Biol. 59, 519 (2008). DOI:10.1146/annurev.arplant.59.032607.092839 [CrossRef] [PubMed] [Google Scholar]
- A. Genre, G. Russo, Front. Plant Sci. 7 (2016). DOI:10.3389/fpls.2016.00096 [Google Scholar]
- L.H. Madsen, L. Tirichine, A. Jurkiewicz, J.T. Sullivan, A.B. Heckmann, A.S. Bek, C.W. Ronson, E.K. James, J. Stougaard, Nat. Commun. 1, 10 (2010). DOI:10.1038/ncomms1009 [PubMed] [Google Scholar]
- A.Y. Borisov, T.N. Danilova, T.A. Koroleva, E.V. Kuznetsova, L. Madsen, M. Mofett, T.S. Naumkina, T.A. Nemankin, E.S. Ovchinnikova, Z.B. Pavlova, N.E. Petrova, A.G. Pinaev, S. Radutoiu, S.M. Rozov, T.S. Rychagova, O.Y. Shtark, I.I. Solovov, J. Stougaard, I.A. Tikhonovich, A.F. Topunov, V.E. Tsyganov, A.G. Vasil’chikov, V.A. Voroshilova, N.F. Weeden, A.I. Zhernakov, V.A. Zhukov, Appl. Biochem. Microbiol. 43, 237 (2007) [Google Scholar]
- V.E. Tsyganov, V.A. Voroshilova, U.B. Priefer, A.Y. Borisov, I.A. Tikhonovich, Ann. Bot. 89, 357 (2002). DOI:10.1093/aob/mcf051 [CrossRef] [PubMed] [Google Scholar]
- V.A. Voroshilova, B. Boesten, V.E. Tsyganov, A.Y. Borisov, I.A. Tikhonovich, U.B. Priefer, Mol. Plant Microbe Interact. 14, 471 (2001). DOI:10.1094/MPMI.2001.14.4.471 [CrossRef] [PubMed] [Google Scholar]
- A.M. Hirsch, New Phytol. 122 (1992). DOI:10.1111/j.1469-8137.1992.tb04227.x [Google Scholar]
- A.C.J. Timmers, M.C. Auriac, G. Truchet, Development 126, 3617 (1999) [PubMed] [Google Scholar]
- J. Dénarié, F. Debellé, J.-C. Promé, Annu. Rev. Biochem. 65, 503 (1996). DOI:10.1146/annurev.bi.65.070196.002443 [CrossRef] [PubMed] [Google Scholar]
- F.C. Guinel, R.D. Geil, Can. J. Bot. 80, 695 (2002). DOI:10.1139/b02-066 [Google Scholar]
- S. Radutoiu, L.H. Madsen, E.B. Madsen, A. Jurkiewicz, E. Fukai, E.M.H. Quistgaard, A.S. Albrektsen, E.K. James, S. Thirup, J. Stougaard, EMBO J. 26, 3923 (2007). DOI:10.1038/sj.emboj.7601826 [CrossRef] [PubMed] [Google Scholar]
- A.N. Kirienko, Y.B. Porozov, N.V. Malkov, G.A. Akhtemova, C. Le Signor, R. Thompson, C. Saffray, M. Dalmais, A. Bendahmane, I.A. Tikhonovich, E.A. Dolgikh, Planta 248, 1101 (2018). DOI:10.1007/s00425-018-2944-4 [CrossRef] [PubMed] [Google Scholar]
- S. Gonzalez-Rizzo, M. Crespi, F. Frugier, Plant Cell. 18, 2680 (2006). DOI:10.1105/tpc.106.043778 [Google Scholar]
- P.H. Middleton, J. Jakab, R.V. Penmetsa, C.G. Starker, J. Doll, P. Kalo, R. Prabhu, J.F. Marsh, R.M. Mitra, A. Kereszt, B. Dudas, K. VandenBosch, S.R. Long, D.R. Cook, G.B. Kiss, G.E.D. Oldroyd, Plant Cell. 19, 1221 (2007). DOI:10.1105/tpc.106.048264 [Google Scholar]
- P. Smit, J. Raedts, V. Portyanko, F. Debellé, C. Gough, T. Bisseling, R. Geurts, Science 308, 1789 (2005). DOI:10.1126/science.1111025 [Google Scholar]
- X. Liu, R. Wolfe, L.R. Welch, D.S. Domozych, Z.A. Popper, A.M. Showalter, PLoS One 11 (2016). DOI:10.1371/journal.pone.0150177 [Google Scholar]
- R. Mirabella, C. Franken, G.N.M. Van Der Krogt, T. Bisseling, R. Geurts, Plant Physiol. 135, 1879 (2004). DOI:10.1104/pp.103.038539 [Google Scholar]
- A. Sevin-Pujol, M. Sicard, C. Rosenberg, M.C. Auriac, A. Lepage, A. Niebel, C. Gough, S. Bensmihen, PLoS One 12 (2017). DOI:10.1371/journal.pone.0188923 [Google Scholar]
- A.N. Kirienko, N.A. Vishnevskaya, A.B. Kitaeva, O.Y. Shtark, P.Y. Kozyulina, R. Thompson, M. Dalmais, A. Bendahmane, I.A. Tikhonovich, E.A. Dolgikh, Int. J. Mol. Sci. 20, 1624 (2019). DOI:10.3390/ijms20071624 [Google Scholar]
- M. Karimi, B. De Meyer, P. Hilson, Trends Plant Sci. 10, 103 (2005). DOI:10.1016/j.tplants.2005.01.008 [CrossRef] [PubMed] [Google Scholar]
- I.V. Leppyanen, A.N. Kirienko, E.A. Dolgikh, Peer J. 7 (2019). DOI:10.7717/peerj.6552 [Google Scholar]
- V.A. Voroshilova, K.N. Demchenko, N.J. Brewin, A.Y. Borisov, I.A. Tikhonovich, New Phytol. 181, 913 (2009). DOI:10.1111/j.1469-8137.2008.02723.x [CrossRef] [PubMed] [Google Scholar]
- P. Rival, F. de Billy, J.J. Bono, C. Gough, C. Rosenberg, S. Bensmihen, Development. 139, 3383 (2012). DOI:10.1242/dev.081620 [CrossRef] [PubMed] [Google Scholar]
- S. Bensmihen, F. de Billy, C. Gough, PLoS One 6, e26114 (2011). DOI:10.1371/journal.pone.0026114 [CrossRef] [PubMed] [Google Scholar]
- S. Moling, A. Pietraszewska-Bogiel, M. Postma, E. Fedorova, M.A. Hink, E. Limpens, T.W.J. Gadella, T. Bisseling, Plant Cell 26, 4188 (2014). DOI:10.1105/tpc.114.129502 [Google Scholar]
- G.E.D. Oldroyd, J.D. Murray, P.S. Poole, J.A. Downie, Annu. Rev. Genet. 45, 119 (2011) [CrossRef] [PubMed] [Google Scholar]
All Figures
 |
Fig. 1 Scheme of pKm43GW p35S::K1::t35S (a) and pKm43GW pLeEXT1::K1::t35S (b) vectors for pea plant transformation. |
| In the text | |
 |
Fig. 2 Nodules on transgenic roots of k1-1 pea mutant line transformed with vectors a) p35S::K1::t35S b) pLeEXT1::K1::t35S. Plants were inoculated with GUS-labeled R. leguminosarum bv. viciae CIAM1026. Scale bar – 0, 3 mm. |
| In the text | |
 |
Fig. 3 The number of nodules and primordia on the transgenic roots of the k1-1 line transformed with the p35S::K1::t35S and pLeEXT1::K1::t35S constructs during inoculation with GUS-labelled R. leguminosarum bv. viciae strain. The data of 4 independent biological repeats were analyzed. The Mann-Whitney U-test was used to estimate the differences between two variants. The differences between variants can be considered as insignificant (using significance level p = 0.05). |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.



