| Issue |
BIO Web Conf.
Volume 30, 2021
II International Symposium “Innovations in Life Sciences” (ILS 2020)
|
|
|---|---|---|
| Article Number | 05005 | |
| Number of page(s) | 6 | |
| Section | Fundamental and Applied Aspects of Biotechnology and Microbiology | |
| DOI | https://doi.org/10.1051/bioconf/20213005005 | |
| Published online | 22 April 2021 | |
Biotechnological potential of fungi and bacteria with ligninolytic activity (mini-review)
Belgorod State National Research University, Belgorod, Russia
* Corresponding author: solyanikova@bsu.edu.ru
The presented mini-review gives a general idea of oxidative enzymes of fungi and bacteria. Possible directions of their practical application are shown.
© The Authors, published by EDP Sciences, 2021
 This is an Open Access article distributed under the terms of the Creative Commons Attribution License 4.0, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License 4.0, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
1 Introduction
A huge amount of pollutants enters the environment as a result of industrial development, urbanization and intensive agriculture with the simultaneous use of a large number of pesticides. In recent years, more and more attention has been paid to the development of and the Introduction of resource-saving technologies and increasing the degree of environmental safety of industrial enterprises through the further use of various natural nutrient substrates. Lignin, the second after cellulose naturally occurring compound, is a highly crosslinked polymer formed by the polymerization of 4hydroxyphenylpropanoid monomers (monolignols) via ether and carbon-carbon bonds. Phenolic fragments of monomer units are p-hydroxyphenyl, guaiacyl and syringyl groups, and the percentage of them depends on the type of plant and tissue (Fig. 1).
Lignin can be considered not only as a pollutant of paramount importance. In an expanding bioprocessing industry, in which large quantities of carbohydrate biomass are subsequently used to produce chemicals, further valorization of lignin is an essential part of the processing process. Depolymerization of lignin leads to the formation of a mixture of monomeric and oligomeric aromatic compounds. The use and conversion of these lignin-based aromatic compounds will pave the way to get various products in biorefineries [1, 2]. Lignocellulosic plant wastes from agriculture and the woodworking industry are the dominant type of potential plant material for microbiological conversion. When using biological agents, plant waste can be processed into valuable target products while reducing the rate of environmental pollution.
The ability of microorganisms to destroy the lignocellulosic complex formed from plant waste is not the same in terms of the duration and processing depth. One of the promising biological agents producing ligninolytic enzymes are microand macroscopic fungi [3]. As for bacteria, in recent years the number of publications showing a high potential of this group of microorganisms in the destruction of ligno-cellulose residues has increased significantly. The purpose of this work is to evaluate the possibility of usage bacteria and fungi in biotechnologies for cleaning the environment from ligno-cellulose residues.
2 Material and Methods
The main materials of this review were obtained as a result of a study of literary sources.
3 Results and discussion
Due to chemical bonds and many aromatic residues, lignin is compound highly resistant to a degradation [4]. The lignin degradation enzymes of white rot fungi are most fully studied [5]. Lignin destruction is initiated by peroxidases and / or laccases. Under the action of peroxidases on lignin, its oxidative fragmentation occurs with the formation of polyphenolic products. The formation of dimers occurs during the oxidation of phenolic monolignols to their corresponding phenolic radicals. Along with this, there are processes of condensation of lignin. Reverse polymerization is facilitated by subsequent enzymatically-catalyzed oneelectron oxidations [6, 7]. Condensed lignin containing C—C diphenyl structures exhibits high resistance to enzymes, which leads to the formation of chains of dark granules, and this is the reason for the appearance of a “marble” pattern on wood [8]. There is an opinion that peroxidase has a wider substrate spectrum than other enzymes of the lignolytic complex [9]. It has been established that they use various types of hemecontaining peroxidases, which include the so-called lignin peroxidases (LiP), manganese peroxidases (MnP), universal peroxidases (VP) and peroxidases, bleaching dyes (DyP) [10]. Lignin peroxidase (EC 1.11.1.14) is a heme-containing glycoprotein that catalyzes the cleavage of C–C bonds in polymer lignin molecules, the oxidation of benzyl alcohols and methyl groups in benzyl compounds, and the opening of an aromatic ring [11]. Manganese peroxidase (EC 1.11.1.13) is a glycoprotein containing heme as a cofactor. The enzyme oxidizes Mn2+ ions to Mn3+ in the presence of hydrogen peroxide, which, in turn, oxidize phenol derivatives, hydroxylate aromatic compounds [12].
The lignolytic complex of basidiomycetes also includes phenol oxidase laccase (paradiphenol: oxygen oxidoreductase, EC 1.10.3.2). Laccase is a coppercontaining glycoprotein that catalyzes the oxidation of polyphenols and polyamines, some inorganic ions in the reaction of reduction of molecular oxygen to water [13]. Laccases are capable of catalyzing the one-electron oxidation of organic compounds to the corresponding radical species. Laccases in the catalytic center contain a cluster of 4 copper ions, oxygen serves as an electron acceptor, with the formation of water as a by-product. The resulting radical products may undergo further oxidation, hydration, disproportionation, or polymerization. The work of the enzyme leads to the formation of phenoxy radicals and quinones, which initiate spontaneous reactions, including the breaking of C–C bonds in aromatic substrates, their demethoxylation, demethylation and polymerization, as well as the formation of reactive oxygen species. Unlike ligninolytic peroxidases, which are characteristic exclusively of white rot basidiomycetes and their litter fungi, laccase is also produced by ascomycetes, deuteromycetes, bacteria and is the most common oxidative enzyme in soils [14].
Information about the catalytic properties of fungal laccases, their isoenzyme composition is known, but only for limited genera [15]. The most thoroughly studied are the oxidoreductases of basidiomycetes [16], while the literature on the deuteromycete enzymes is still contradictory and the number of fungi studied in this direction is small. It is also known that the vital activity of imperfect fungi is crucial for the initial stages of the biodestructive process and the preparation of material components for utilization by other groups of destructive organisms. It was established that the maximum activity of Aspergillus terreus and Trichoderma viride peroxidase, phenol oxidase, and catalase in most cases is observed on days 10–13, while the pH of the medium optimal for the manifestation of enzyme activity is in the range 7.2–8.2 [17]. Micromycetes are natural active destructors of wood lignin; phenol oxidase of microscopic fungi from the genera Trichoderma and Fusarium are involved in biodegradation of lignin [18, 19].
It is known that microscopic fungi, in comparison with bacteria, exhibit the highest exoenzymatic activity and use up to 60% of the substrate to build their cells, while in bacteria its utilization rate is on average 35% [20]. It should be noted that only xylotrophic fungi are able to decompose lignin, possessing a complex of oxidase and hydrolase enzymes. The decomposition of all components of the lignocellulosic substrate can occur almost simultaneously or with the predominant decomposition of lignin, while the cellulose remains relatively unaffected, which depends on the sequence of induction of certain enzymes [21].
Intensive delignification of hardwood is accompanied by the active consumption of xylose, which is part of the glucuronoxylan hemicellulose complex. To decompose lignin, a cosubstrate is needed, which some hemicelluloses can serve. A characteristic of the destructive ability of xylotrophic fungi is the loss of wood caused by them in the laboratory. For 196 days, the fungus may be able to destroy spruce wood by 54%, and aspen by 58%. The decomposition of lignin by xylotrophic fungi is directly proportional to the loss of wood mass and has a linear relationship [22].
It is known that it is advisable to search for promising strains for the destruction of plant residues among soil micromycetes [23]. Thus, strains of Trichoderma koningii, Ulocladium botrytis, Curvularia lunata, Alternaria alternata, and Bipolaris australiensis with high values of lignin degradation ability were isolated from the rhizosphere of leguminous plants [23, 24].
In contrast to the relatively well-studied fungal ligninolytic complex, the ligninolytic ability of bacteria still need to be studied. In recent years, a growing number of articles describes bacterial lignolytic enzymes. Typical lignin peroxidases characteristic of fungi were not found in bacteria. Perhaps this is due to the peculiarities of the expression of these rather complex glycosylated proteins containing several disulfide bonds and including several calcium ions and a heme cofactor [6]. However, studies have shown that bacteria possess a specific type of peroxidase called bleach dyes peroxidase (Dy-P) [25]. The first Dy-P was described in 1999, but only recently it has been shown that this enzyme is widely distributed in bacterial genomes [10]. Dy-P was found in Escherichia coli K-12, Thermobifida fusca YX, Rhodococcus jostii RHA1, Streptomyces viridosporus strain T7A, Streptomyces coelicolor A, Amycolatopsis sp. 75iv2, Pseudomonas sp. strain YS-1p.
Depending on the sequence, Dy-Ps in the database are divided into four classes [26]. Enzymes belonging to classes AC are mainly found in bacteria, while members of the class D-DyPs are extracellular representatives of fungi [27]. Class A Dy-Ps typically have a Tat signal sequence, i.e. are secreted. The protein sequences of class B and C Dy-Ps are intracellular enzymes. The InterPro database currently contains 8318 Dy-P sequences. But only about thirty of them were isolated and characterized [27, 28]. Dy-Ps are mainly active at acidic pH and exhibit a very broad substrate profile, including several classes of synthetic dyes, monophenol compounds, veratril alcohol, β-carotenoids, Mn2+ and lignin model compounds, but their physiological substrates remain unknown. In general, the bacterial DyPs studied to date have a lower oxidative ability than fungal analogues, are usually active with monophenol substrates, but some bacterial Dy-Ps show significant activity against non-phenolic veratril alcohol. Some bacterial Dy-Ps are capable of oxidizing 4methoxymandelate in the presence of Mn2+ and O2, without requiring the presence of H2O2, acting as oxidases. Very promising is the Dy-P of strain Pseudomonas fluorescence Pf-5, capable of modifying alkaline kraft lignin, a by-product of paper production. This is the most stable bacterial peroxidase (60% residual activity after incubation at 70 ° C for 2 h) with high stability in alkaline solutions (> 80% activity after incubation at pH 5-10 at 37 ° C for 1 h) [29].
The second type of ligninolytic enzymes is lignin-modifying laccases, including bacterial ones. Like fungal laccases, many bacterial laccases are secreted and usually carry out polymerization or depolymerization reactions. The Tat secretion system is used to export bacterial laccases. Since laccases do not need cofactors and do not form toxic H2O2, they are biotechnologically significant biocatalysts, unlike many oxidoreductases. The greatest number of laccases is characterized in representatives of the genus Streptomyces. Laccases are also known in bacteria of the genera Bacillus and Thermus. Laccases are used in bioremediation processes for disinfecting industrial wastewater, in the food industry for stabilizing drinks and improving the organoleptic properties of food products, in the synthesis of pharmaceuticals and other fine chemicals, in the manufacture of textile dyes and in the delignification of wood, especially in the bleaching process [30]. Laccases can also be used in pretreatment of coniferous species to improve subsequent hydrolysis. Laccases exhibit high potential in biotransformation of various pollutants. These enzymes are used as crude extracts of enzymes, free or immobilized enzymes. Application of laccase in bioremediation and water treatment on a large scale are limited by the complex composition and high salt concentration and pH values of contaminated media that affect protein stability, recovery, and utilization. These problems also associated with operational problems and the need for large-scale production of varnished products [31]. Therefore, based on data on the characteristics of water bodies that need to introduce treatment technologies, it is necessary to identify and develop new laccases that can be used in difficult conditions and develop new strategies and processes to achieve their effective application in the treatment of contaminated water. Recently, the problems of stability, efficiency, separation and reuse have overcome the immobilization of enzymes and the development of new biocatalytic materials.
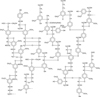 |
Fig. 1. The structure of lignin |
Microbial strain capable of lignin degradation and their biotechnology application
4 Conclusion
For the successful use of lignolytic enzymes in industry, of particular interest are more stable enzymes with high catalytic activity. The search for such enzymes is of the great interes. Three strategies can be mentioned for the search for microorganisms with the ability to decompose lignin. The first one is the isolation, characterization and verification of each individual strain. In the Table the most perspective strains are listed. As an example, gene encoding a laccase from a thermophilic aerobic bacteria Caldalkalibacillus thermarum TA2.A1 was cloned and expressed in E. coli [32]. It was found that laccase from this strain was monomeric protein with a molecular weight of 57 kDa.
The second approach is based on the emergence and development of new biotechnologies and bioinformatics tools that allow the search for new enzymes using the metagenome. These approaches can make a significant contribution to the future economical production from renewable resources [39]. The third approach is the creation of highly efficient multiple analysis and strain search systems with specified characteristics, performed by the newly created enzyme systems. One of such highthroughput screening strategy for searching alkali lignindegrading microorganisms developed by Chong et al. [40] is based on the ferricyanide-based assay involving the formation of Prussian blue. The use of this screening system made it possible to isolate from soil the strain Rhodococcus pyridinivorans CCZU-B16, capable of degradation 30.2% of 4 g/L alkali lignin under the nitrogen-limited condition at 30 °C for 72 h. In addition, a distinctive feature of this strain was the accumulation of lipids in the cells. Among the C16 and C18 fatty acid profiles, four major ones prevailed, including palmitic acid, palmitoleic acid, stearic acid and oleic acid [40].
Based on the data obtained by various scientists studying the ability of microorganisms to transform lignin, we can conclude the high potential significance of such microorganisms for different areas of biotechnology, e.g. to clean the environment of lignin, alkali lignin resulting from the pulp and paper industry, for processing plant residues of agricultural waste, to obtain high-value compounds from lignin. In light of the data obtained by various scientists studying the ability of microorganisms to transform lignin, we can conclude the high potential significance of such microorganisms for different areas of biotechnology, e.g. pulp and paper industry, textile, food industries, bioremediation, organic synthesis, pharmaceutical sector, nanobiotechnology, a biofuel cell, and processing of agricultural waste [41].
Acknowledge
This work was carried out in the framework of the program Scientific and Educational Center “Innovative solutions in the agro-industrial complex”, Belgorod. The authors thank the staff of the Department of Biotechnology and Microbiology and students A.S. Barskovoi, E.P. Yesina and Yu.V. Gushchina for technical assistance in conducting a series of laboratory experiments.
References
- O.Y. Abdelaziz, D.P. Brink, J. Prothmann, K. Ravi, M. Sun, J. García-Hidalgo, M. Sandahl, C.P. Hulteberg, C. Turner, G. Lidén, M.F. Gorwa Grauslund, Biotechnol. Adv. 34(8), 1318 (2016) [PubMed] [Google Scholar]
- A.J. Ragauskas, G.T. Beckham, M.J. Biddy, R. Chandra, F. Chen, M.F. Davis, B.H. Davison, R.A. Dixon, P. Gilna, M. Keller, P. Langan, A.K. Naskar, J.N. Saddler, T.J. Tschaplinski, G.A. Tuskan, C.E. Wyman, Science 344(6185), 1246843, (2014) [PubMed] [Google Scholar]
- T.V. Ryazanova, N.A. Chuprova, T.A. Luneva, in Catal. Ind. 6, 64 (2014) [Google Scholar]
- A.M. Abdel-Hamid, J.O. Solbiati, I.K.O. Cann, Adv. Appl. Microbiol. 82, 1 (2013) [PubMed] [Google Scholar]
- E.V. Dorshakova, N.P. Elinov, A.N. Mamoshin, Problems of Medical Mycology 16(4), 41 (2014) [Google Scholar]
- G. de Gonzalo, D.I. Colpa, M.H.M. Habib, M.W. Fraaij, J. Biotechnol. 236, 110 (2016) [PubMed] [Google Scholar]
- P. Muradov, Sh. Gasymov, F. Gahramanova, A. Alieva, D. Abbasova, Sh. Babaeva, M. Ragimova, Vestnik MGOU, 1, 109–112 (2009). [Google Scholar]
- G.N. Kononov, A.N. Veryovkin, N.L. Goryachev, S.S. Nikitin, Lesnoi vestnik 6, 112 (2015) [Google Scholar]
- V.V. Shutova, V.V. Revin, and T.V. Kudaeva, in Isvestia of Saratov University, part Chemistry, Biology, Ecology, 13, 80–85 (2015). [Google Scholar]
- C. Lambertz, S. Ece, R. Fischer, U. Commandeur, Bioengineered, 7, 145 (2016) [PubMed] [Google Scholar]
- S. Sarkanen, R.A. Razal, T. Piccariello, E. Yamamoto, N.G. Lewis, J. Biol. Chem. 266(6), 3636 (1991) [PubMed] [Google Scholar]
- E. Karhunen, A. Kantelinen, M.-L. Niku-Paavola, Arch. Biochem. Biophys., 279(1), 25 (1990) [PubMed] [Google Scholar]
- A.I. Yaropolov, O.V. Skorobogat’ko, S.S. Vartanov, S.D. Varfolomeyev, Appl. Biochem. Biotechnol 49(3), 257 (1994) [Google Scholar]
- A.G. Zavarzina, T.A. Semenova, O.V. Belova, A.V. Lisov, A.A. Leontievskii, A.E. Ivanova, Microbiology, 87(3), 233 (2018) [Google Scholar]
- L. Kh. Khalimova, L.I. Zakirova, N.I. Petukhova, V.V. Zorin, Bashkirian Chemical Journal, 18(4), 166 (2011) [Google Scholar]
- T.V. Fedorova, N.V. Shakhova, O.I. Klein, O.A. Glazunova, L.G. Maloshenok, N.A. Kulikova, N.V. Psurtseva, O.V. Koroleva Appl. Biochem. Microbiol. 49(6), 570 (2013) [Google Scholar]
- E.S. Kasatova “The activity of exooxidoreductases of microscopic fungi in connection with their biodegradation of natural and synthetic polymers“: avtoreferat, N. Novgorod, 2011. [Google Scholar]
- I.A. Gneusheva, N.E. Pavlovskaya, I.V. Yakovleva, Vestnik OrelSAU 3(24), 36 (2010) [Google Scholar]
- I.V. Darmov, E.I. Gorshunova, T.S. Tarasova, Scientific notes of the Kazan University. Natural Sciences Series, 159(1), 72 (2017) [Google Scholar]
- I.S. Sandanova, “Microbiological destruction of plant litter of steppe ecosystems of southeastern Transbaikalia”: research, 2007: 127. [Google Scholar]
- I.A. Kazartsev, V.I. Roshchin, VA. Solovyov, Mycology and phytopathology 48(2), 112 (2014) [Google Scholar]
- K.A. Vyrastkova, I.G. Wide Adv. Sci. 1, 7 (2017) [Google Scholar]
- Yu.N. Kurkina, A.S. Barskova, and E.P. Esina, “Study of kinetics of micromycetes, which produced lignolytic enzymes in vitro” in “Current status, problems and prospects for the development of agricultural science: materials of the IV International Scientific and Practical Conference, Yalta, sept. 2019. [Google Scholar]
- Yu.N. Kurkina, Vegetables crops of Russia, 6, 114 (2019) [Google Scholar]
- E. van Bloois, D.E. Torres Pazmi˜no, R.T. Winter, M.W. Fraaije Appl. Microbiol. Biotechnol. 86, 1419 (2010) [PubMed] [Google Scholar]
- N. Fawal, Nucleic Acids Res. 41, 441 (2013) [Google Scholar]
- T. Yoshida, Y. Sugano, Arch. Biochem. Biophys. 574, 49 (2015) [PubMed] [Google Scholar]
- D.I. Colpa, M.W. Fraaije, E. van Bloois, J. Ind. Microbiol. Biotechnol. 41, 1 (2014) [PubMed] [Google Scholar]
- W. Yu, W. Liu, H. Huang, F. Zheng, X. Wang, Y. Wu, K. Li, X. Xie, Y. Jin, PLoS One, 9, e110319 (2014) [PubMed] [Google Scholar]
- R.S. Shraddha, S. Sehgal, M. Kamthania, A. Kumar, Enzyme Res. Article ID 217861. [Google Scholar]
- L. Arregui, M. Ayala, X. Gómez Gil, G. Gutiérrez Soto, C.E. Hernández Luna, M.H. de los Santos, L. Levin, A. Rojo Domínguez, D. Romero Martínez, M.C.N. Saparrat, M.A. Trujillo Roldán, N.A. Valdez Cruz, Microb Cell Fact. 18, 200 (2019) [PubMed] [Google Scholar]
- S. Ghatge, Y. Yang, W.-Y. Song, T.-Y Kim, H.-G. Hur Appl. Microbiol. Biotechnol. 102, 4075 (2018) [PubMed] [Google Scholar]
- K. T. Steffen, A. Hatakka, M. Hofrichterm, Appl. Environm. Microbiol. 68(7), 3442 (2002) [Google Scholar]
- Y. Yanagi, K. Kitayama, H. Tahara, N. Fujitake, “Decolorization of soil fulvic acids by laccases from Trametes versicolor and Trametes villosa [Google Scholar]
- E.S. Lasareva, V.F. Smirnov, I.V. Struchkova, Bulletin of the Nizhny Novgorod University. N.I. Lobachevsky 1, 77 (2008) [Google Scholar]
- O.V. Turkovskaya, N.N. Pozdnyakova, Bulletin of the Ufa Scientific Center of the Russian Academy of Sciences 3(5), 60 (2018) [Google Scholar]
- A.A. Klepikov, M.M. Shamtsyan, Izvestiya SPbGTI (TU) 3, 39 (2014) [Google Scholar]
- A.K. Chandel, O.V. Singh, Appl. Microbiol. Biotechnol. 89, 1289 (2011) [PubMed] [Google Scholar]
- T.H. Al-kahem Al-balawi, A.L. Wood, A. Solis, T. Cooper, R.D. Barabote, Curr Microbiol. 74, 762 (2017) [PubMed] [Google Scholar]
- G.-G. Chong, X. J. Huang, J. H. Di, D. Z. Xu, Y.C. He, Y.N. Pei, Y.J. Tang, C.L. Ma, Bioprocess. Biosyst. Eng. 41, 501 (2018) [PubMed] [Google Scholar]
- A. Kunamneni, F. J. Plou, A. Ballesteros, M. Alcalde, Recent Pat Biotechnol 2(1) 10 (2008) [PubMed] [Google Scholar]
All Tables
Microbial strain capable of lignin degradation and their biotechnology application
All Figures
 |
Fig. 1. The structure of lignin |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.



