| Issue |
BIO Web Conf.
Volume 68, 2023
44th World Congress of Vine and Wine
|
|
|---|---|---|
| Article Number | 01018 | |
| Number of page(s) | 6 | |
| Section | Viticulture | |
| DOI | https://doi.org/10.1051/bioconf/20236801018 | |
| Published online | 06 December 2023 | |
Phenobot - Intelligent photonics for molecular phenotyping in Precision Viticulture
1 INESC TEC - Institute for Systems and Computer Engineering, Technology and Science, Porto, Portugal
2 FCUP - Department of Geosciences, Environment and Spatial Planning, Faculty of Sciences of the University of Porto, Portugal
3 INIAV - National Institute of Agricultural and Veterinary Research, I.P., Portugal
4 ADVID - Associação para o Desenvolvimento da Viticultura Duriense (ADVID) - CoLAB Vines&Wines – National Collaborative Laboratory for the Portuguese Wine Sector. Vila Real, Portugal
5 COTHN - National Horticultural Operative and Technological Center, Alcobaça, Portugal
6 SPIN.WORKS - Av. da Igreja 42 6º, 1700-239 Lisboa, Portugal
7 Portugal Ramos - Adega Vila Santa, Estrada Nacional 4, 7100-149 Estremoz, Portugal
8 Francisco Rodrigues - Quinta do Monte Bravo, 5130-130 Ervedosa do Douro, Portugal
9 Poças - Sociedade Vinícola Terras de Valdigem, S.A, Peso da Régua, Portugal
10 Real Companhia Velha - Rua de Azevedo Magalhães, 314, 4430-022 Vila Nova de Gaia, Portugal
11 Frutus - Estação Fruteira de Montejunto, CRL, Portugal
12 Camoptec - Comercialização e Consultoria em Hortofrutícolas, Portugal
13 Quinta do Pinto Sociedade Agrícola e Comercial, S.A. – Aldeia Galega da Merceana, Portugal
* Corresponding author: rui.c.martins@inesctec.pt
The Phenobot platform is comprised by an autonomous robot, instrumentation, artificial intelligence, and digital twin diagnosis at the molecular level, marking the transition from pure data-driven to knowledge-driven agriculture 4.0, towards a physiology-based approach to precision viticulture. Such is achieved by measuring the plant metabolome ‘in vivo' and 'in situ', using spectroscopy and artificial intelligence for quantifying metabolites, e.g.: i. grapes: chlorophylls a and b, pheophytins a and b, anthocyanins, carotenoids, malic and tartaric acids, glucose and fructose; ii. foliage: chlorophylls a and b, pheophytins a and b, anthocyanins, carotenoids, nitrogen, phosphorous, potassium, sugars, and leaf water potential; and iii. soil nutrients (NPK). The geo-referenced metabolic information of each plant (organs and tissues) is the basis of multi-scaled analysis: i. geo-referenced metabolic maps of vineyards at the macroscopic field level, and ii. genome-scale 'in-silico' digital twin model for inferential physiology (phenotype state) and omics diagnosis at the molecular and cellular levels (transcription, enzyme efficiency, and metabolic fluxes). Genome-scale 'in-silico' Vitis vinifera numerical network relationships and fluxes comprise the scientific knowledge about the plant's physiological response to external stimuli, being the comparable mechanisms between laboratory and field experimentation - providing a causal and interpretable relationship to a complex system subjected to external spurious interactions (e.g., soil, climate, and ecosystem) scrambling pure data-driven approaches. This new approach identifies the molecular and cellular targets for managing plant physiology under different stress conditions, enabling new sustainable agricultural practices and bridging agriculture with plant biotechnology, towards faster innovations (e.g. biostimulants, anti-microbial compounds/mechanisms, nutrition, and water management). Phenobot is a project under the Portuguese emblematic initiative in Agriculture 4.0, part of the Recovery and Resilience Plan (Ref. PRR: 190 Ref. 09/C05-i03/2021 – PRR-C05-i03-I-000134).
© The Authors, published by EDP Sciences, 2023
 This is an Open Access article distributed under the terms of the Creative Commons Attribution License 4.0, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License 4.0, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
1 Introduction
Current Precision Viticulture (PV) relies heavily on data science and large-scale sensing methods such as multispectral sensors, drones, satellites, and individual instruments for measuring moisture and temperature. This approach does not prioritize the use of in-situ physiology-based diagnostics supported by Omics measurements. Managing the complex interactions between genotype, environment, and management (GEM) using a systematic, quantitative, and causal methodology is currently challenging with these approaches. The primary obstacle is the lack of tools for real-time, in-situ monitoring of plant metabolism, which are still in the research phase and have not been widely implemented or made available in the context of Agriculture 4.0.
PV relies heavily on data, falling short in representing the intricate soil-plant-climate system on a mechanistic basis. Consequently, when environmental conditions deviate from historical data, models are unable to accurately predict plant metabolic and physiological networks. Additionally, data-intensive PV techniques are not easily transferable between different locations or years, since perturbations on relationships are influenced by various complex factors acting at different scales. Relying solely on data-based AI approaches instead of considering the plant's molecular and physiological mechanisms is a significant obstacle in PV's development. This challenge has hindered the effective implementation of biotechnological strategies in advanced PV and needs a shift to mechanistic cause-effect approaches to provide reliable information on plant physiological diagnosis.
The Future of PV faces significant challenges, such as how to: i) implement targeted and precise practices based on plant physiology; ii) use advanced high-throughput digital sensors; and iii) integrate these sensors with systems biology in silico to explore grapevine metabolic pathways and gain a better understanding on how field physiology and metabolism are driven by Genotype-Environment-Management interactions. INESC TEC Omics robotic platforms, such as Metbots, represent the cutting-edge in ‘in-situ’ metabolomics, leveraging advanced Point-of-Measurement (POM) spectroscopy analysis to quantify i) leaves (pigments, minerals, metabolites, and biomolecules), ii) fruits (including pigments, sugars, organic acids, metabolites, and biomolecules), and iii) sap: sucrose, glucose, fructose, and NPK.
Here in we present the Phenobots project - a robotic platform for Intelligent photonics for high-throughput ‘in-vivo’, ‘in-situ’ and ‘in-silico’ vine molecular phenotyping in PV, comprising of: i. autonomous robot for geo-referenced individual plant monitoring (soil, leaf, fruits); ii. Point-of-Measurement system for metabolomics, physiology state, and precision fertilization; iii. Tomography-like metabolomic internal structures assessment and multi-scaled diagnosis; and iv. inferential genome-scale models digital twins for performing inferential omics diagnosis.
2 Autonomous robotic platform
There are currently few robots available for use in agriculture. Although some robotic solutions have been developed for specific agricultural tasks over the last two decades, many of these solutions are not easily scalable or replicable to other farms due to variations in terrain and crop types. However, INESCTEC's robotics lab has developed specialized robotic platforms, such as AgRob V16 and ROMOVI (P2020 project), designed to operate on steep slopes in the Douro Valley terrain. These platforms have overcome challenges related to GPS signal issues, harsh terrain conditions that limit instrumentation, and slopes that require precise path planning.
2.1 Sensors and navigation
The present generation of robots uses our developed Vineyard Simultaneous Localization and Mapping VineSLAM system [1], taking into consideration the natural and artificial features of the vineyard to recognize the localization, which compensates for poor GPS accuracy. Tests of AgRob V16 in a real steep slope that this platform can overcome ditches, rocks, and high slopes (30%). With a robust localization system, it can perform autonomously a crop monitoring task (crop yield, soil/air temperature/humidity, and crop water stress index), being cost-effective for the end-user.
The system is divided in three main layers:
Perception: 3D point cloud processing to extract edge, planar and semiplane features;
Mapping: Multimodal registration of the types of features extracted to build a consistent 3D map of the agriculture environment;
Localization: PF-based procedure that uses both point- and semiplane-based information to localize the robot.
Thus, our approach can efficiently extract point and semi plane features from a 3D point cloud and use them to build a map of the crop, localizing the robot within this map (Fig. 1) [2].
 |
Figure 1 Autonomous robotic platform: (a) robot; (b) VineSLAM navigation system reconstructed virtual vineyard map ([2]). |
2.2 Vision system and Robotic Arm Measurement
Measurement of the vine leaf and grapes is performed by the POM mounted on the robotic arm (Fig. 2a). After positioning the robot in front of the desired vine plant, the following procedure is performed: i. using machine vision to locate vine leaves and grape bunches (Fig. 2b) [3]; ii. pin-point the grape or leaf to be measured [3-5]; iii. calculate the best arm trajectory until 0.5 cm proximity; iv. trajectory control with camera and proximity sensor [6,],; and v. use spectrometer data to finetune the approach in the final 0.5 cm for performing the measurement. Grape and leaves recognition using deep learning models work as follows:
Data collection: video data recorded by cameras mounted on top of an agricultural robotic platform; image extraction and storage from videos to build the input dataset;
Dataset generation: image annotation by drawing bounding boxes around grape bunches in images considering two different classes; image augmentation by the application of several operations to the images and annotations to increase the dataset size and avoid overfitting when training the DL models; image splitting of the image size, to avoid losing resolution due to the image resize operation performed by the models to their kernel size (in this case, 300 × 300 px, with three channels);
Model training and deployment: training and quantization of the DL models to deploy them in a low-cost and low-power embedded device with the main goal of performing time-effective grape bunch detection in images (Fig. 2) [3].
 |
Figure 2 Autonomous robotic platform: (a) robotic arm with POM, camera, and proximity sensor, (b) computer vision deep-learning model for leaf and grape/grape bunch recognition; and (c) path planning for performing geo-referenced metabolic mapping using spectroscopy. |
3 Point-of-measurement system
3.1 Hardware and software
The POM system is a UV-Vis-SWNIR (200-1200 nm) Intelligent of things (IoT) system, using INESC TEC AgIoT platform and dedicated spectroscopy electronics [8-10] (Fig. 3). The IoT capacity allows the device to be remotely operated without any software installation, so that is immediately plugged into the robot arm, or used as a stand-alone portable intelligent photonics laboratory (Fig. 3a) [10,11]. The POM can simultaneously measure solid structures (e.g. leaf, grapes, soil) or liquids (sap, fertilizer, grape juice, fermentation) by using a plug-in capsule system as described in [12]. The POM has no other than an on/off key and is an intelligent device system that can use distributed learning system for creating a collaborative big database for the spectroscopy AI system [13].
An example of ease of usage is the direct comparison of measuring the water potential: i. Scholander chamber (Igor Gonçalves - ADVID, Portugal) vs. ii. Metbots POM (Renan Tosin - INESC TEC, Portugal). Using the Sholander chamber is extremely laborious and not practical, whereas the POM is less than 150g and the leaf water potential is measured and estimated in less than 1s [11] (Fig. 3a).
 |
Figure 3 Point-of-measurement intelligent photonics laboratory: (a) manual hydric potential using Scholander chamber vs using Metbots POM device; (b) POM with capsule system and IoT Software on mobile phone; (c) direct quantification of glucose, fructose, malic and tartaric acids in several grape varieties [10]. |
3.2 Spectroscopy information processing
Spectroscopy data in complex samples, such as plant structures (e.g., leaves or grapes) is characterized by multi-scaled interference and matrix effects, carrying both physical and chemical information in a single spectral. Unscrambling the existing information contained in each spectrum lies at the heart of intelligent photonics, allowing ‘in-vivo’ metabolic quantification and a key enabling technology for physiology-based viticulture.
Spectral information is pre-processed for removing scattering artifacts and afterward subjected to processing by i. Feature space optimization: information about a constituent is present in the spectra in different scales and wavelengths; and ii. Covariance mode (CovM) search: searching a group of samples within the feature space that belong to the same interference pattern, that is, the gradient mixture of interferents, where information is proportional to the constituent concentration correct features and transforms [10,11]. The developed state-of-the-art CovM search method allows for a significant decrease in the error estimation, allowing to attain significantly lower detection limits and several metabolites possible to be quantified from a single spectrum, such as i. leaves (chlorophylls a/b, pheophytins a/b, anthocyanins, carotenoids, nitrogen, potassium, phosphorous, water potential); ii. grapes (chlorophylls a/b, pheophytins a/b, anthocyanins, carotenoids, glucose, fructose, malic and tartaric acid, total acidity, tannins, nitrogen) [10,11]; iii. sap nutrition (NPK); or iv. fertilizer (NPK) [14] (Fig. 3.).
4 Tomography-like metabolomics
The POM capacities have recently been updated into providing internal tissue reconstruction with the SpecTOM technology [15]. The optical configuration of the POM allows it to record internal tissue information as described in Fig. 4. The observed recorded spectra are the convolution of the internal tissue spectra of the different spectra: skin, pulp, and seed (Fig. 4). An innovative method using hierarchical latent spaces modelling is used to provide a bi-directional relationship between the observed spectra to the different tissues spectra, allowing to infer the internal tissue spectra once a grape spectrum is observed (Fig. 4), which enables to determine the maturation stage of each tissue or to detect maturation abnormalities in internal tissues (e.g. seeds).
 |
Figure 4 Grape internal tissue decomposition for direct analysis of skin, pulp, and seed metabolomics. |
5 Genome-scale model
Genome-scale models (GEMs) (Fig. 5) are state-of-the-art mechanistic descriptions of all existing annotated genome-scale relationships between genes, proteins, enzymes, and metabolites, representing metabolic pathways, regulation mechanisms, and cell transport [16]. This knowledgebase is translated into a numerical model, representing the calculation of pseudo-steady states given by the known physiological constraints (e.g., protons, nutrients, CO2, photon intensity, and quantified omics data) to define a phenotype space and determine physiological cell states [17-19]. Current models largely benefited from Bioinformatics and Systems Biology infrastructure for genome, proteome, expression/regulation, and pathways annotation, allowing to assemble full plant ‘omics’ model with multi-organ and tissue capacities [20-22]. For example, tomato multi-organ GEMs (root, stem, and leaves) can accurately predict metabolic fluxes in the whole plant for central metabolism, the impact of photon intensity on photosynthesis in leaves and stems, sap composition in xylem and phloem, and the impact of nitrate (NO3) deficiency on growth rate [22].
Gap knowledge challenging (grapevine) GEM physiology-based PV, includes: i) incomplete gene annotation; ii) limited knowledge of metabolic pathways that difficult accurate modelling of the metabolism of grapevine cells; iii) lack of experimental data (e.g. gene expression data, metabolite measurements, and enzyme activities); and iv) limited integration with other omics data to fully understand the metabolic network of the grapevine.
The Phenobot project is currently building a Vitis vinifera GEM with multi-organ capabilities (Fig. 5), based on the duration of VitisNet [23] in curated and annotated genes, transcripts, transporters, enzymes, and metabolites (7854 genes and transcripts, 1631 proteins, and 1998 metabolites), integrating the root, stem, grape, and leaves with xylem/phloem transport by interconnecting each organ through input-output metabolites and nutrients (Fig. 5) [22].
The GEM model is used for both simulation and diagnosis. Simulation is performed using constrained-based flux balance analysis (FBA) using climacteric data (temperature, light intensity, and cycle) (Fig. 2). Flux constraints are set from both experimental and bibliographic data. Diagnosis is performed by constrain-based FBA with regression against ‘greenhouse’ controlled experiments ‘omics’ quantification - using transcription, proteomics, and metabolomics results to characterize and diagnose gene-protein-enzyme-flux of the functional state within the grapevine phenotype space [24,25] (Fig. 5). The GEM model is used to infer metabolic fluxes and active pathways, where gene transcription and enzyme efficiency is given by external variables (nutrients and climate). GEMs can link the information between the field and biotechnology laboratory through the analysis of the expressed phenotype and by understanding the limitations of the phenotype space under different conditions, which can either limit or enhance cell functions. Phenotype spaceallows targeting specific cellular mechanisms, introducing viticulture into a more quantitative and scientific approach towards physiology-based Precision Viticulture.
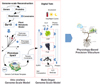 |
Figure 5 'In-Silico' Genome scale modelling for physiology-based Precision Viticulture. |
6 Time-course monitoring
Time-course monitoring of individual plants’ physiology can be analyzed at the vineyard level, by the creation of time-course and geo-referenced spatiotemporal maps (Figs. 6 and 7). Previous results in Douro Valley to Touriga Nacional, show that there is a direct relationship between fruit maturation, terroir features, and climate conditions; and that these are extremely dynamic.
(Figs. 6 and 7). Previous results in Douro Valley to Touriga Nacional, show that there is a direct relationship between fruit maturation, terroir features, and climate conditions; and that these are extremely dynamic.
For example, key moments in grape maturation lead to increased metabolic variability and dependence on terroir and climate, such as during the veraison and harvest periods. (Fig. 6) shows that greater differences in maturation occur at these stages and that the terrain water retention is a great discriminant of the spectral characteristics of grapes in terms of pigments, sugar, and acids. When climacteric conditions are more stable and grape development is within veraison-harvest, grape composition tends to be more uniform along the vineyard (Fig. 6).
In another example, one can observe that the grape maturation index (ratio sugars/acids) evolves at a slow rate until the 19th of August, and after this point, a significant acceleration in maturation is observed (Fig. 7).
The acceleration in maturation is also highly correlated to the terroir features, where grapes monitored on the left-hand side generally present higher maturation ratios of sugars/acids. Phenobots will revise this data with GEMs to try to understand what changes are occurring at the molecular level, triggering these very significant differences in physiological response to climacteric and terroir during grape maturation.
 |
Figure 6 Grape geo-referenced metabolic maps from verasion to harvest. |
 |
Figure 7 Grape geo-referenced variance maps from verasion to harvest. |
7 Conclusions
The technology presented in this manuscript is the continuous effort to develop state-of-the-art tools that can enable ‘in-vivo’ molecular ‘omics’ diagnosis for implementing physiology-based precision viticulture. Furthermore, it is expected that GEMs can link plant biotechnology and field research, for implementing targeted molecular management of plant physiology, a new way of performing Precision Agriculture.
This research was funded by Pheonobot - Intelligent Photonics for Phenotyping of Agro-Food Crops (09/C05-i03/2021 – PRR-C05-i03-I-000134) and OmicBots: High-Throughput Integrative Omic-Robots Platform for a Next Generation Physiology-based Precision Viticulture (PTDC/ASP-HOR/1338/2021). Silva FM, Tosin R and Pereira MR acknowledge Fundação para a Ciência e Tecnologia (FCT) PhD research grants Ref. DFA/BD/9136/2020, SFRH/BD/145182/2019 and SFRD/BD/146564/2019. Martins RC acknowledges Fundação para a Ciência e Tecnologia (FCT) research contract grant (CEEIND/017801/2018).
References
- F.N. dos Santos, H. Sobreira, D. Campos, R. Morais, A. Moreira, O. Contente, J of Intelligent & Robotic Sys. 83(3-4), 429-444 (2016) [CrossRef] [Google Scholar]
- A.S. Aguiar, F. Neves dos Santos, H. Sobreiro, J. Boaventura-Cunha, A.J. Sousa, Front. Robot. AI 9, 832165 (2022) [CrossRef] [Google Scholar]
- A.S. Aguiar, S.A. Magalhães, F.N. dos Santos, L. Castro, T. Pinho, J. Valente, R.C. Martins, J. Boaventura-Cunha, Agron J. 11, 1890 (2021) [Google Scholar]
- R. Berenstein, O.B. Shahar, A. Shapiro, Y. Edan, Intell. Serv. Robot. 3(4), 233–243 (2010) [CrossRef] [Google Scholar]
- S.S. Mehta, T.F. Burks, Comput Electron Agric 102, 146-158(2014) [CrossRef] [Google Scholar]
- Y.K. Hwang, N. Ahuja, ACM Comput. Surv. 24(3), 219–291 (1992) [CrossRef] [Google Scholar]
- Y. Mezouar, F. IEEE Trans. Robot. 18(4), 534–549 (2002) [CrossRef] [Google Scholar]
- T.G. Barroso, L. Ribeiro, H. Gregório, F. Santos, R.C. Martins, Sens. Actuators B Chem. 343, 130138 (2021) [CrossRef] [Google Scholar]
- T.G. Barroso, L. Ribeiro, H. Gregório, F. Monteiro-Silva, F. Neves dos Santos, R.C. Martins, Chemosensors 10, 460 (2022) [CrossRef] [Google Scholar]
- R.C. Martins, T.G. Barroso, P. Jorge, M. Cunha, F. Santos, Comput Electron Agric. 194, 106710 (2022) [CrossRef] [Google Scholar]
- R. Tosin, R.C. Martins, I. Poças, M. Cunha. Biosyst. Eng. 219, 235-258 (2022) [CrossRef] [Google Scholar]
- US10209178B2 Optical system for parameter characterization of an element of body fluid or tissue. [Google Scholar]
- EP19838971 A calibration method of a spectroscopy device comprising a plurality of sensors and of transfer of spectral information obtained from at least two calibrated spectroscopy devices [Google Scholar]
- F. Monteiro-Silva, P.A.S. Jorge, R.C. Martins, Chemosensors 7(4), 51, 2019 [CrossRef] [Google Scholar]
- PT115801 Method and device for providing non-invasive tomographic characterization of a sample comprising a plurality of differentiated tissues [Google Scholar]
- B.O. Palsson, Systems Biology - Constraint-based reconstruction and analysis. Cambridge University Press, Cambridge, United Kingdom (2015) [CrossRef] [Google Scholar]
- I. Famili, R. Mahadevan, B.O. Palsson, Biophys. J. 88(3), 1616–1625 (2005) [CrossRef] [Google Scholar]
- N.D. Price, J.L. Reed, J.A. Papin., I. Famili, B.O. Palsson, Biophys. J. 84(2), 794–804 (2003) [CrossRef] [Google Scholar]
- Y. Xi, F. Wang, PLoS One 14(2), e0210539 (2019) [Google Scholar]
- L. Gerlin, C. Frainay, F. Jourdan, C. Baroukh, S. Prigent, S. Plant genome-scale metabolic networks. In P. Pétriacq & A. Bouchereau (Eds.), Advances in Botanical Research 98 (237-270) Academic Press. (2021) [CrossRef] [Google Scholar]
- C.G. de Oliveira Dal'Molin, L.E. Quek, P.A. Saa, L.K. Nielsen, Front. Plant Sci. 6, 4 (2015) [Google Scholar]
- L. Gerlin, L. Cottret, A. Escourrou, S. Genin, C. Baroukh, Plant Physiol. 188(3), 1709–1723 (2022) [CrossRef] [PubMed] [Google Scholar]
- J. Grimplet, G.R. Cramer, J.A. Dickerson, K. Mathiason, J. Van Hemert, A.Y. Fennell, PLoS One 4(12), e8365 (2009) [CrossRef] [PubMed] [Google Scholar]
- M.L. Mo, M.L.B.Ø. Palsson, M.J. Herrgård, BMC Syst. Biol. 3, 37 (2009) [CrossRef] [Google Scholar]
- N. Lewis, H. Nagarajan, B. Palsson, Nat. Rev. Microbiol. 10, 291-305 (2012) [CrossRef] [PubMed] [Google Scholar]
All Figures
 |
Figure 1 Autonomous robotic platform: (a) robot; (b) VineSLAM navigation system reconstructed virtual vineyard map ([2]). |
| In the text | |
 |
Figure 2 Autonomous robotic platform: (a) robotic arm with POM, camera, and proximity sensor, (b) computer vision deep-learning model for leaf and grape/grape bunch recognition; and (c) path planning for performing geo-referenced metabolic mapping using spectroscopy. |
| In the text | |
 |
Figure 3 Point-of-measurement intelligent photonics laboratory: (a) manual hydric potential using Scholander chamber vs using Metbots POM device; (b) POM with capsule system and IoT Software on mobile phone; (c) direct quantification of glucose, fructose, malic and tartaric acids in several grape varieties [10]. |
| In the text | |
 |
Figure 4 Grape internal tissue decomposition for direct analysis of skin, pulp, and seed metabolomics. |
| In the text | |
 |
Figure 5 'In-Silico' Genome scale modelling for physiology-based Precision Viticulture. |
| In the text | |
 |
Figure 6 Grape geo-referenced metabolic maps from verasion to harvest. |
| In the text | |
 |
Figure 7 Grape geo-referenced variance maps from verasion to harvest. |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.



